HDF5 in Igor Pro
HDF5 is a widely-used, very flexible data file format. The HDF5 file format is a creation of the National Center for Supercomputing Applications (NCSA). HDF5 is now supported by The HDF Group (http://www.hdfgroup.org).
Igor Pro includes deep support for HDF5, including:
-
Browsing HDF5 files
-
Loading data from HDF5 files
-
Saving data to HDF5 files
-
Saving Igor experiments as HDF5 files
-
Loading Igor experiments from HDF5 files
Prior to Igor Pro 9.00, HDF5 support was provided by an XOP that you had to activate. HDF5 support is now built into Igor and activation is no longer required.
Support for saving and loading Igor experiments as HDF5 files was added in Igor Pro 9.00. For details see HDF5 Packed Experiment Files.
Index of HDF5 Topics
Here are the main sections in the following material on HDF5:
HDF5 Guided Tour
This section will get you off on the right foot using HDF5 in Igor Pro.
In the following material you will see numbered steps that you should perform. Please perform them exactly as written so you stay in sync with the guided tour.
This tour in intended to help experienced Igor users learn how to access HDF5 files from Igor but also to entice non-Igor users to buy Igor. Therefore the tour is written assuming no knowledge of Igor.
HDF5 Overview
HDF5 is a very powerful but complex file format that is designed to be capable of storing almost any imaginable set of data and to encapsulate relationships between data sets.
An HDF5 file can contain within it a hierarchy similar to the hierarchy of directories and files on your hard disk. In HDF5, the hierarchy consists of "groups" and "datasets". There is a root group named "/". Each group can contain datasets and other groups.
An HDF5 dataset is a one-dimensional or multi-dimensional set of elements. Each element can be an "atomic" datatype (e.g., 16-bit signed integer or a 32-bit IEEE float) or a "composite" datatype such as a structure or an array. A "compound" datatype is a composite datatype similar to a C structure. Its members can be atomic datatypes or composite datatypes. For now, forget about composite datatypes - we will deal with atomic datatypes only.
Each dataset can have associated with it any number of "attributes". Attributes are like datasets but are attached to datasets, or to groups, rather than being part of the hierarchy.
Igor Pro HDF5 Support
The Igor Pro HDF5 package consists of built-in HDF5 support and a set of Igor procedures.
The Igor procedures, which are automatically loaded when Igor is launched, implement an HDF5 browser in Igor. The browser supports:
-
Previewing HDF5 file data
-
Loading HDF5 datasets and groups into Igor
-
Saving Igor waves and data folders in HDF5 files
In Igor Pro 9 and later, Igor can save Igor experiments as HDF5 packed experiment files and reload experiments from them.
Using the HDF5 Browser
-
Choose Data→Load Waves→New HDF5 Browser.
This displays an HDF5 browser control panel.
As you can see, the HDF5 Browser takes up a bit of screen space. You will need to arrange it and this help window so you can see both.
-
Click the Open HDF5 File button and open the following file:
Igor Pro Folder\Examples\Feature Demos\HDF5 Samples\TOVSB1NF.h5(If you're not sure where your Igor Pro Folder is, choose Misc→Path Status, click on the Igor symbolic path, and note the path to the Igor Pro Folder.)
We got this sample from the NCSA web site.
You should now see something like this:
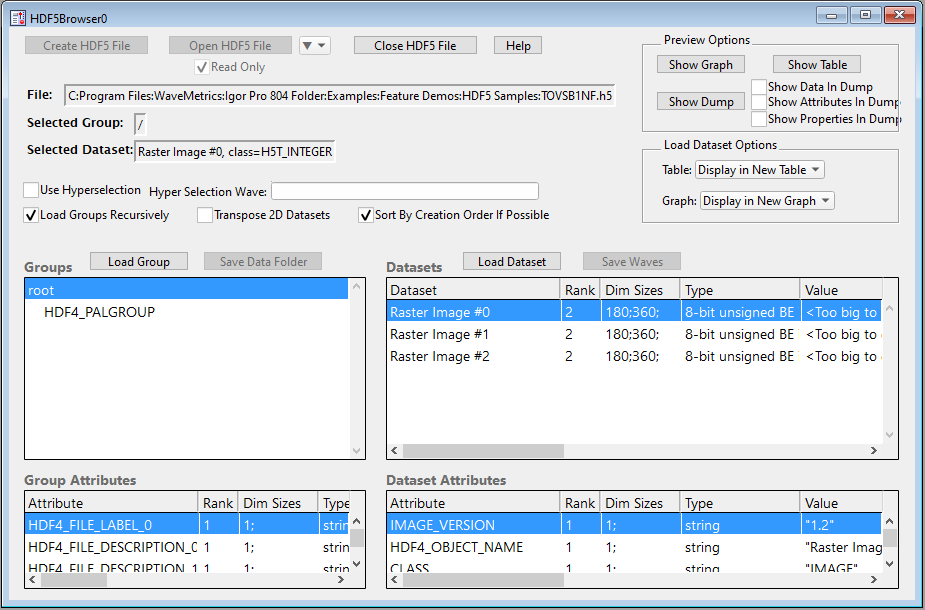
The browser contains four lists.
The top/left list is the Groups list and shows the groups in the HDF5 file. Groups in an HDF5 file are analogous to directories in a hard disk hierarchy. In this case there are two groups, root (which is called "/" in HDF5 terminology) and HDF4_PALGROUP. HDF4_PALGROUP is a subgroup of root.
This file contains a number of objects with names that begin with HDF4 because it was created by converting an HDF4 file to HDF5 format using a utility supplied by The HDF Group.
Below the Groups list is the Group Attributes list. In the picture above, the root group is selected so the Group Attributes list shows the attributes of the root group. An attribute is like a dataset but is attached to a group or dataset instead of being part of the HDF5 file hierarchy. Attributes are usually used to save small snippets of information related to a group or dataset.
The top/right list is the Datasets list. This lists the datasets in the selected group, root in this case. In the root group of this file we have three datasets all of which are images.
Below the Datasets list is the Dataset Attributes list. It shows the attributes of the selected dataset, Raster Image #0 in this case.
Three of the lists have columns that show information about the items in the list.
-
Familiarize yourself with the information listed in the columns of the lists.
To see all the information you will need to either scroll the list and/or resize the entire HDF5 browser window to make it larger.
-
In the Groups list, click the subgroup and notice that the information displayed in the other lists changes.
-
In the Groups list, click the root group again.
Now we will see how to browse a dataset.
-
Click the Show Graph, Show Table and Show Dump buttons and arrange the three resulting windows so that they can all be seen at least partially.
These browser preview windows should typically be kept fairly small as they are intended just to provide a preview. It is usually convenient to position them to the right of the HDF5 browser.
The three windows are blank now. They display something only when you click on a dataset or attribute.
-
In the Datasets list, click the top dataset (Raster Image #0).
The dataset is displayed in each of the three preview windows.
The dump window shows the contents of the HDF5 file in "Data Description Language" (DDL). This is useful for experts who want to see the format details of a particular group, dataset or attribute. The dump window will be of no interest in most everyday use.
If you check the Show Data in Dump checkbox and then click a very large dataset, it will take a very long time to dump the data into the dump window. Therefore you should avoid checking the Show Data in Dump checkbox.
The preview graph and table, not surprisingly, allow you to preview the dataset in graphical and tabular form.
This dataset is a special case. It is an image formatted according to the HDF5 Image and Palette Specification which requires that the image have certain attributes that describe it. You can see these attributes in the Dataset Attributes list. They are named CLASS, IMAGE_VERSION, IMAGE_SUBCLASS and PALETTE. The HDF5 Browser uses the information in these attributes to make a nice preview graph.
An HDF5 file can contain a 2D dataset without the dataset being formatted according to the HDF5 Image and Palette Specification. In fact, most HDF5 files do not follow that specification. We use the term "formal image" to make clear that a particular dataset is formatted according to the HDF5 Image and Palette Specification and to distinguish it from other 2D datasets which may be considered to be images.
-
In the Dataset Attributes list, click the CLASS attribute.
The value of the selected attribute is displayed in the preview windows.
Try clicking the other image attributes, IMAGE_VERSION, IMAGE_SUBCLASS and PALETTE.
So far we have just previewed data, we have not loaded it into Igor. (Actually, it was loaded into Igor and stored in the root:Packages:HDF5Browser data folder, but that is an HDF5 Browser implementation detail.)
Now we will load the data into Igor for real.
-
Make sure that the Raster Image #0 dataset is selected, that the Table popup menu is set to Display In New Table and that the Graph popup menu is set to Display In New Graph. Then click the Load Dataset button.
The HDF5 Browser loads the dataset (and its associated palette, because this is a formal image with an associated palette dataset) into the current data folder in Igor and creates a new graph and a new table.
-
Choose Data→Data Browser and note the two "waves" in the root data folder.
"Wave" is short for "waveform" and is our term for a dataset. This terminology stems from our roots in time series signal processing.
The two waves, 'Raster Image #0' and 'Raster Image #0Pal' were loaded when you clicked the Load Dataset button. The graph was set up to display 'Raster Image #0' using 'Raster Image #0Pal' as a palette wave.
-
Back in the HDF5 Browser, with the root group still selected, click the Load Group button.
The HDF5 Browser created a new data folder named TOVSB1NF and loaded the contents of the HDF5 root group into the new data folder which can be seen in the Data Browser. The name TOVSB1NF comes from the name of the HDF5 file whose root group we just loaded.
When you load a group, the HDF5 Browser does not display the loaded data in a graph or table. That is done only when you click Load Dataset and also depends on the Load Dataset Options controls.
-
Click the Close HDF5 File button and then click the close box on the HDF5 Browser.
If you had closed the HDF5 browser without clicking the Close HDF5 File button, the browser would have closed the file anyway. It will also automatically close the file if you choose File→New Experiment or File→Open Experiment or if you quit Igor.
Next we will learn how to write an Igor procedure to access an HDF5 file programmatically. Before you start that, feel free to play with the HDF5 Browser on your own HDF5 files. Start by choosing Data→Load Waves→New HDF5 Browser.
Igor can handle most HDF5 files that you will encounter, but it cannot handle all possible HDF5 files. If you receive an error while examining your own files, it may be because of a bug or because Igor does not support a feature used in your file. In this case you can send the file along with a brief explanation of the problem to support@wavemetrics.com and we will determine whether the problem is a bug or just a limitation.
Loading HDF5 Data Programmatically
Igor includes a number of operations and functions for accessing HDF5 files. To get an idea of the range and scope of the operations, click the link below. Then return here to continue the guided tour.
In this section, we will load an HDF5 dataset using a user-defined function.
-
Choose File→New Experiment.
You will be asked if you want to save the current experiment. Click No (or Yes if you want to save your current work environment).
This closes the current experiment, killing any waves and windows.
-
Choose Misc→New Path and create an Igor symbolic path named HDF5Samples and pointing to the HDF5 Samples directory which contains the TOVSB1NF.h5 file that we used in the preceding section.
The New Path dialog will look something like this:
Click Do It to create the symbolic path.
An Igor symbolic path is a short name that references a specific directory on disk. We will use this symbolic path in a command that opens an HDF5 file.
In HDF5, you must first open a file or create a new file. The HDF5 library returns a file ID that you use in subsequent calls to the library. When you are finished, you must close the file. The following example illustrates this three step process.
-
Choose Windows→Procedure Window and paste the following into the Procedure window:
Function TestLoadDataset(datasetName)
String datasetName // Name of dataset to be loaded
Variable fileID // HDF5 file ID will be stored here
Variable result = 0 // 0 means no error
// Open the HDF5 file
HDF5OpenFile /P=HDF5Samples /R /Z fileID as "TOVSB1NF.h5"
if (V_flag != 0)
Print "HDF5OpenFile failed"
return -1
endif
// Load the HDF5 dataset
HDF5LoadData /O /Z fileID, datasetName
if (V_flag != 0)
Print "HDF5LoadData failed"
result = -1
endif
// Close the HDF5 file
HDF5CloseFile fileID
return result
EndRead through the procedure. Notice that we use the symbolic path HDF5Samples created above as a parameter to the HDF5OpenFile operation.
The /R flag was used to open the file for read-only so that we don't get an error if the file cannot be opened for writing.
The /Z flag tells HDF5OpenFile that, if there is an error, it should set the variable V_flag to a non-zero value but return to Igor as if no error occurred. This allows us to handle the error gracefully rather than having Igor abort execution and display an error dialog.
HDF5OpenFile sets the local variable fileID to a value returned from the HDF5 library. We pass that value to the HDF5LoadData operation. We also provide the name of a dataset within that file to be loaded. Again we check the V_flag variable to see if an error occurred.
Finally we call HDF5CloseFile to close the file that we opened. It is important to always close files that we open. If, during development of a procedure, you forget to close a file or fail to close a file because of an error, you can close all open HDF5 files by executing:
HDF5CloseFile /A 0Also, Igor automatically closes all open files if you choose File→New Experiment, File→Revert Experiment or File→Open Experiment or if you quit Igor.
-
Click the Compile button at the bottom of the Procedure window.
If you don't see a Compile button at the bottom of the Procedure window, that is because the procedures were already automatically compiled when you deactivated the Procedure window.
If you get a compile error then you have not pasted the right text into the Procedure window or you have entered other incorrect text. Try going back to step 1 of this section.
Now we are ready to run our procedure.
-
Execute the following command in the Igor command line either by typing it and pressing Enter, by copy/paste followed by Enter, or by selecting it and pressing Ctrl-Enter:
TestLoadDataset("Raster Image #0")At this point the procedure should have correctly executed and you should see no error messages printed in the history area (just above the command line). This history area of the command window should contain:
Waves loaded by HDF5LoadData: Raster Image #0
Now we will verify that the data was loaded.
-
Choose Data→Data Browser. Then double-click the wave named 'Raster Image #0'.
This creates a table showing the data just loaded.
At this point Igor's root data folder contains just one wave, 'Raster Image #0', and does not contain the corresponding palette wave. That's because we loaded the dataset as a plain dataset, not as a formal image. The HDF5LoadData operation does not know about formal images. For that we need to use HDF5LoadImage.
-
In the Data Browser, right-click the 'Raster Image #0' wave and choose New Image.
Igor creates an image plot of the wave.
Because we loaded 'Raster Image #0' as a plain dataset, not as a formal image, the image plot is gray instead of colored. Also, it is rotated 90 degrees relative to the image plot we saw earlier in the tour. That's because Igor plots 2D data different from most programs and the HDF5LoadData operation does not compensate by default.
Our procedure is of rather limited value because it is hard-coded to use a specific symbolic path and a specific file name. We will now make it more general.
Before we do that, we will save the current work environment so that, if you make a mistake, you can revert to a version of it that worked.
-
Choose File→Save Experiment As and save the current work environment in a new Igor experiment file named "HDF5 Tour.pxp" in a directory where you store your own files.
This saves all of your data and procedures in a single file. If need be, later you can revert to the saved state by choosing File→Revert Experiment.
-
Open the Procedure window (Windows→Procedure Window) and replace the first two lines of the TestLoadDataset function with this:
Function TestLoadDataset(pathName, fileName, datasetName)
String pathName // Name of symbolic path
String fileName // Name of HDF5 file
String datasetName // Name of dataset to be loaded -
Change the HDF5OpenFile command to this:
HDF5OpenFile /P=$pathName /R /Z fileID as fileNameHere we replaced HDF5Samples with $pathName. $pathName tells Igor to use the contents of the string parameter pathName as the parameter for the /P flag. We also replaced "TOVSB1NF.h5" with fileName so that we can specify the file to be loaded when we call the function instead of when we code it.
-
Click the Compile button and then execute this in the command line:
TestLoadDataset("HDF5Samples", "TOVSB1NF.h5", "Raster Image #0")This does the same thing as the earlier version of the function but now we have a more general function that can be used on any file in any directory.
The command above reloaded the 'Raster Image #0' dataset into Igor, overwriting the previous contents. Since the new contents is identical to the previous contents, the graph and table did not change.
-
Choose File→Save Experiment to save your current work environment in the HDF5 Tour.pxp experiment file that you created earlier.
You can now take a break and quit Igor if you want.
Saving HDF5 Data Programmatically
Now that we have seen how to programmatically load HDF5 data we will turn our attention to saving Igor data in an HDF5 file.
In this section, we will create some Igor data and save it in an HDF5 dataset from a user-defined function.
-
If the HDF5 Tour.pxp experiment that you previously saved is not already open, open it by choosing File→Recent Experiments→HDF5 Tour.pxp.
Next we will create some data that we can save in an HDF5 file. We will create the data in a new Igor data folder to keep it separate from our other data.
-
Choose Data→Data Browser. Click the New Data Folder button. Enter the name Test Data, click the Set As Current Data Folder and click OK.
The Data Browser shows the new data folder and a red arrow pointing to it. The red arrow indicates the current data folder. Operations that do not explicitly address a specific data folder work in the current data folder.
-
On Igor's command line, execute these commands:
Make/N=100 data0, data1, data2
SetScale x 0, 2*PI, data0, data1, data2
data0=sin(x); data1=2*sin(x+PI/6); data2=3*sin(x+PI/4)
Display data0, data1, data2The SetScale operation set the X scaling for each wave to run from 0 to 2π. The symbol x in the wave assignment statements takes on the X value for each point in the destination wave as the assignment is executed.
(If you have not already done it, later you should do the Igor guided tour by choosing Help→Getting Started. It explains X scaling and wave assignment statements as well as many other Igor features.)
-
Choose Misc→New Path and create an Igor symbolic path named HDF5Data pointing to a directory on your hard disk where you will save data.
Choose Misc→Path Status and verify that the HDF5Data symbolic path exists and points to the intended directory.
Now that we have done some more work worth saving we will save the current experiment so we can revert to a known good state if necessary.
-
Choose File→Save Experiment to save your current work environment in the HDF5 Tour.pxp experiment file that you created earlier.
If need be, later you can revert to the saved state by choosing File→Revert Experiment.
-
Choose Windows→Procedure Window and paste the following into the Procedure window below the TestLoadDataset function:
Function TestSaveDataset(pathName, fileName, w)
String pathName // Name of symbolic path
String fileName // Name of HDF5 file
Wave w // The wave to be saved
Variable result = 0 // 0 means no error
Variable fileID
// Create a new HDF5 file, overwriting if same-named file exists
HDF5CreateFile/P=$pathName /O /Z fileID as fileName
if (V_flag != 0)
Print "HDF5CreateFile failed"
return -1
endif
// Save wave as dataset
HDF5SaveData /O /Z w, fileID
if (V_flag != 0)
Print "HDF5SaveData failed"
result = -1
endif
// Close the HDF5 file
HDF5CloseFile fileID
return result
EndRead through the procedure. It should look familiar as it is similar to the TestLoadDataset function we wrote before. Again it has parameters that specify a symbolic path and file name. It does not have a parameter to specify the dataset name because HDF5SaveData uses the wave name as the dataset name unless instructed otherwise. This function has a wave parameter through which we will specify the wave whose data is to be written to the HDF5 file.
Here we use HDF5CreateFile to create a new file rather than HDF5OpenFile. HDF5CreateFile creates a new file and opens it, returning a file ID. If you wanted to add a dataset to an existing file you would use HDF5OpenFile instead of HDF5CreateFile.
HDF5CreateFile sets the local variable fileID to a value returned from the HDF5 library. We pass that value to the HDF5SaveData operation. The /O flag means that, if there is already a dataset with the same name, it will be overwritten.
Finally we call HDF5CloseFile to close the file that we opened via HDF5CreateFile.
-
Click the Compile button at the bottom of the Procedure window.
If you get an error then you have not pasted the right text into the Procedure window or you have entered other incorrect text. If you cannot find the error, choose File→Revert Experiment and go back to step 6 of this section.
Now we are ready to run our procedure.
-
Execute the following command in the Igor command line either by typing it and pressing Enter, by copy/paste followed by Enter, or by selecting it and pressing Ctrl-Enter:
TestSaveDataset("HDF5Data", "SaveTest.h5", data0)At this point the procedure should have correctly executed and you should see no error messages printed in the history area of the command window (just above the command line).
Now we will verify that the data was saved.
-
Choose Data→Load Waves→New HDF5 Browser. Click the Open HDF5 File button and open the SaveTest.h5 file that we just created.
Verify that the file contains a dataset named data0.
The data0 dataset has some attributes. These attributes allow HDF5LoadData to fully recreate the wave and all of its properties if you ever load it back into Igor. If you don't want to save these attributes you can use the /IGOR=0 flag when calling HDF5SaveData.
-
Click the Close HDF5 File button.
We can't write more data to the file while it is open in the HDF5 Browser.
The TestSaveDataset function is rather limited because it saves just one wave. We will now make it more general so that it can save any number of waves. But first we will save our work since it is in a known good state.
-
Choose File→Save Experiment to save your current work environment in the HDF5 Tour.pxp experiment file that you created earlier.
If need be, later you can revert to the saved state by choosing File→Revert Experiment.
Next we will create a new, more general user function with a different name (TestSaveDatasets instead of TestSaveDataset).
-
Choose Windows→Procedure Window and paste the following into the Procedure window below the TestSaveDataset function:
Function TestSaveDatasets(pathName, fileName, listOfWaves)
String pathName // Name of symbolic path
String fileName // Name of HDF5 file
String listOfWaves // Semicolon-separated list of waves
Variable result = 0 // 0 means no error
Variable fileID
// Create a new HDF5 file, overwriting if same-named file exists
HDF5CreateFile/P=$pathName /O /Z fileID as fileName
if (V_flag != 0)
Print "HDF5CreateFile failed"
return -1
endif
String listItem
Variable index = 0
do
listItem = StringFromList(index, listOfWaves)
if (strlen(listItem) == 0)
break // No more waves
endif
// Create a local reference to the wave
Wave w = $listItem
// Save wave as dataset
HDF5SaveData /O /Z w, fileID
if (V_flag != 0)
Print "HDF5SaveData failed"
result = -1
break
endif
index += 1
while(1)
// Close the HDF5 file
HDF5CloseFile fileID
return result
EndRead through the procedure. It is similar to the TestSaveDataset function.
The first difference is that, instead of passing a wave, we pass a list of wave names in a string parameter. This parameter is a semicolon-separated list which is a commonly-used programming technique in Igor.
The next difference is that we have a do-while loop which extracts a name from the list and saves the corresponding wave as a dataset in the HDF5 file.
The statement
Wave w = $listItemcreates a local "wave reference" and allows us to use w to refer to a wave whose identity is determined by the contents of the listItem string variable. This also is a common Igor programming technique and is explained in detail in the Programming help file.
-
Click the Compile button at the bottom of the Procedure window.
If you get an error then you have not pasted the right text into the Procedure window or you have entered other incorrect text. If you cannot find the error, choose File→Revert Experiment and go back to step 12 of this section.
Now we are ready to run our procedure.
-
Enter the following command in the Igor command line either by typing it and pressing enter, by copy/paste followed by Enter, or by selecting it and pressing Ctrl-Enter:
TestSaveDatasets("HDF5Data", "SaveTest.h5", "data0;data1;data2;")The third parameter is a string in this function. In the previous example, it was a name, specifically the name of a wave. Strings are entered in double-quotes but names are not.
At this point the procedure should have correctly executed and you should see no error messages printed in the history area of the command window (just above the command line).
Now we will verify that the data was saved.
-
Activate the HDF5 Browser window. Click the Open HDF5 File button and open the SaveTest.h5 file that we just created.
Verify that the file contains datasets named data0, data1 and data2.
-
Click the Close HDF5 File button.
-
Choose File→Save Experiment and save the current work environment in the HDF5 Tour.pxp experiment file.
This concludes the HDF5 Guided Tour.
Where To Go From Here
If you are new to Igor or have never done it you should definitely do the Igor Guided Tour. If you are in a hurry, do just the first half of it.
You should also read the following help files which explain the basics of Igor in more detail:
Getting Help, Experiments, Files and Folders, Windows, Waves
To get started with Igor programming you need to read these help sections:
Working With Commands, Programming Overview, User-Defined Functions
For HDF5-specific programming, you need to have at least a basic understanding of HDF5. See the links in the next section. Then you need to familiarize yourself with the HDF5-related operations and functions listed in HDF5 Operations and Functions.
If you run into problems, send a sample HDF5 file along with a description of what you are trying to do to support@wavemetrics.com and we will try to get you started in the right direction.
Learning More About HDF5
In order to use HDF5 operations, you must have at least a basic understanding of HDF5. The HDF5 web site provides an abundance of material. To get started, visit this web page:
https://support.hdfgroup.org/documentation/hdf5/latest/_learn_basics.html
The HDF Group provides a Java-based program called HDFView. You may want to download and install HDFView so that you can easily browse HDF5 files as you read the introductory material. Or you may prefer to use the HDF5 browser provided by Igor.
The HDF5 Browser
Igor Pro includes an automatically-loaded procedure file, "HDF5 Browser.ipf", which implements an HDF5 browser. The browser lets you interactively examine HDF5 files to get an idea of what is in them. It also lets you load HDF5 datasets and groups into Igor and save Igor waves and data folders in HDF5 files.
The browser currently does not support creating attributes. For that you must use the HDF5SaveData operation.
The HDF5 browser includes lists which display:
- All groups in the file
- All attributes of the selected group
- All datasets in the selected group
- All attributes of the selected dataset
In addition, the HDF5 browser optionally displays:
- A graph displaying the selected dataset or attribute
- A table displaying the selected dataset or attribute
- A notebook window containing a dump of the selected group, dataset or attribute
Using The HDF5 Browser
To browse HDF5 files, choose Data→Load Waves→New HDF5 Browser. This creates an HDF5 browser control panel. You can create additional browsers by choosing the same menu item again.
Each browser control panel lets you browse one HDF5 file at a time. For most users, one browser will be sufficient.
After creating a new browser, click the Open HDF5 File button to choose the file to browse.
The HDF5 browser contains four lists which display the groups, group attributes, datasets and dataset attributes in the file being browsed.
HDF5 Browser Basic Controls
Here is a description of the basic controls in the HDF5 browser that most users will use.
Create HDF5 File
Creates a new HDF5 file and opens it for read/write.
Open HDF5 File
Opens an existing HDF5 file for read-only or read/write, depending on the state of the Read Only checkbox.
Close HDF5 File
Closes the HDF5 file.
Show Graph
If you click the Show Graph button, the browser displays a preview graph of subsequent datasets or attributes that you select.
Show Table
If you click the Show Table button, the browser displays a preview table of subsequent datasets or attributes that you select.
Load Dataset
The Load Dataset button loads the currently selected dataset into the current data folder.
Load Dataset Options
This section of the browser contains two popup menus that determine if data that you load by clicking the Load Dataset button is displayed in a table or graph.
The Table popup menu contains three items: No Table, Display in New Table, and Append To Top Table. If you choose Append To Top Table and there are no tables, it acts as if you chose Display in New Table.
The Graph popup menu contains three items: No Graph, Display in New Graph, and Append To Top Graph. If you choose Append To Top Graph and there are no graphs, it acts as if you chose Display in New Graph. Appending is useful when you are loading 1D data but of little use when appending multi-dimensional data. Multi-dimensional data is appended as an image plot which obscures anything that was already in the graph.
Text waves are not displayed in graphs even if Display in New Graph or Append To Top Graph is selected.
Save Waves
Displays a panel that allows you to select and save waves in the HDF5 file, provided the file was open for read/write.
Transpose 2D
If the Transpose 2D checkbox is checked, 2D datasets are transposed to compensate for the difference in how Igor and other programs treat rows and columns in an image plot. See HDF5 Images Versus Igor Images for details. This does not affect the loading of "formal" images (images formatted according to the HDF5 Image and Palette Specification).
Sort By Creation Order If Possible
If checked, and if the file supports listing by creation order, the HDF5 Browser displays and loads groups and datasets in creation order.
Most HDF5 files do not include creation-order information and so are listed and loaded in alphabetical order even if this checkbox is checked. However, HDF5 files written by Igor Pro 9 or later include creation-order information and so can be listed and loaded in creation order.
Load Group
The Load Group button loads all of the datasets in the currently selected group into a new data folder inside current data folder.
The Load Group button calls the HDF5LoadGroup operation using the /IMAG flag. This means that, if the group contains a formal image (see HDF5 Images Versus Igor Images), it is be loaded as a formal image.
The Hyperselection controls do not apply to the operation of the Load Group button.
Load Groups Recursively
If the Load Groups Recursively checkbox is checked, when the Load Group button is pressed any subgroups in the currently selected group are loaded as sub-datafolders.
Load Group Only Once
Because an HDF5 file is a "directed graph" rather than a strict hierarchy, a given group in an HDF5 file can appear in more than one location in the file's hierarchy.
If the Load Group Only Once checkbox is checked, the Load Group button loads a given group only the first time it is encountered. If it is unchecked, the Load Group button loads a given group each time it appears in the file's hierarchy resulting in duplicated data. If in doubt, leave Load Group Only Once checked.
Save Data Folder
Displays a panel that allows you to select a single data folder and save it in the HDF5 file, provided the file was open for read/write.
HDF5 Browser Contextual Menus
If you right-click a row in the Datasets, Dataset Attributes or Group Attributes lists, the browser displays a contextual menu that allows you to perform the following actions:
-
Copy the selected dataset or attribute's value to the clipboard as text
-
Load the selected dataset or attribute as a wave
-
Load all datasets or attributes as a waves
These popup menus also appear if you click the ▼ icons above the lists. They work the same as the HDF5 Browser contextual menus described above.
When the Datasets list is active, choosing Load Selected Dataset as Wave does the same thing as clicking the Load Dataset button.
When copying data to the clipboard, the data is copied as text and consequently may not represent the full precision of the underlying dataset or attribute. Not all datatypes are supported. If the browser supports the datatype that you right clicked, the contextual menu shows "Copy to Clipboard as Text". If the datatype is not supported, it shows "Can't Copy This Data Type".
When loading as waves, any pre-existing waves with the same names are overridden.
When loading waves from datasets, the table and graph options in the Load Dataset Options section of the browser apply. If you check Apply to Attributes also, the options apply when loading waves from attributes.
HDF5 Browser Advanced Controls
Here is a description of the advanced controls in the HDF5 browser. These are for use by people familiar with the HDF5 file format.
Show Dump
If you click the Show Dump button, the browser displays a notebook in which you can see additional details about subsequent groups, datasets or attributes that you select. The dump window is updated each time you select a group, dataset or attribute from any of the lists.
Show Data In Dump
When unchecked, the dump shows header information but not the actual data of a dataset. When checked it shows data as well as header information for a dataset.
If you check the Show Data In Dump checkbox and choose to dump a very large dataset, the dump could take a very long time. If the dump seems to be taking forever, clicking the Abort button in the Igor status bar.
Even if the Show Data In Dump checkbox is checked, the dump for a group consist of the header information only and omits the actual data for datasets and attributes.
Show Attributes In Dump
The Show Attributes In Dump checkbox lets you determine whether attributes are dumped when you select a group or dataset. When checked, information about any attributes associated with the dataset is included in the dump. This checkbox does not affect what is dumped when you select an item in the group or dataset attribute lists.
Show Properties In Dump
The Show Properties In Dump checkbox lets you see properties such as storage layout and filters (compression). This information is usually of little interest but is useful when investigating the effects of compression.
Use Hyperselection
If you check the Use Hyperselection checkbox and enter a path to a "hyperslab wave", the HDF5 Browser uses the hyperselection in the wave to load a subset of subsequent datasets or attributes that you click. This is a feature for advanced users who understand HDF5 hyperselections and have read the HDF5 Dataset Subsets discussion below.
The hyperselection is used when you click the Load Dataset button but not when you click the Load Group button.
HDF5 Browser Dump Technical Details
The dump notebook displays a dump of the selected group, dataset or attribute in "Data Description Language" (DDL) format. For most purposes you will not need the dump window. It is useful for experts who are trying to debug a problem or for people who are trying to understand the nuts and bolts of HDF5.
Sometimes strings in HDF5 files contain a large number of trailing nulls. These are not displayed in the dump window.
Sometimes strings in HDF5 files contain the literal strings "\r", "\n" and "\t" to represent carriage return, linefeed and tab. To improve readability, in the dump window these literal strings are displayed as actual carriage returns, linefeeds and tabs.
HDF5 Operations and Functions
This section lists Igor's HDF5-related operations and functions:
| HDF5CreateFile | Creates a new HDF5 file or overwrites an existing file. | |
| HDF5OpenFile | Opens an HDF5 file, returning a file ID that is passed to other operations and functions. | |
| HDF5CloseFile | Closes an HDF5 file or all open HDF5 files. | |
| HDF5FlushFile | Flushes an HDF5 file or all open HDF5 files. | |
| HDF5CreateGroup | Creates a group in an HDF5 file, returning a group ID that can be passed to other operations and functions. | |
| HDF5OpenGroup | Opens an existing HDF5 group, returning a group ID that can be passed to other operations and functions. | |
| HDF5ListGroup | Lists all objects in a group. | |
| HDF5CloseGroup | Closes an HDF5 group. | |
| HDF5LinkInfo | Returns information about an HDF5 link. | |
| HDF5ListAttributes | Lists all attributes associated with a group, dataset or datatype. | |
| HDF5AttributeInfo | Returns information about an HDF5 attribute. | |
| HDF5DatasetInfo | Returns information about an HDF5 dataset. | |
| HDF5LoadData | Loads data from an HDF5 dataset or attribute into Igor. | |
| HDF5LoadImage | Loads an image written according to the HDF5 Image and Palette Specification version 1.2. | |
| HDF5LoadGroup | Loads an HDF5 group and its datasets into an Igor Pro data folder. | |
| HDF5SaveData | Saves Igor waves in an HDF5 file. | |
| HDF5SaveImage | Saves an image in the format specified by the HDF5 Image and Palette Specification version 1.2. | |
| HDF5SaveGroup | Saves an Igor data folder in an HDF5 group. | |
| HDF5TypeInfo | Returns information about an HDF5 datatype. | |
| HDF5CreateLink | Creates a new hard, soft or external link. | |
| HDF5UnlinkObject | Unlinks an object (a group, dataset, datatype or link) from an HDF5 file. This deletes the object from the file's hierarchy but does not free up the space in the file used by the object. | |
| HDF5DimensionScale | Supports the creation and querying of HDF5 dimension scales. | |
| HDF5LibraryInfo | Returns information about the HDF5 library used by Igor. This is of interest to advanced programmers only. | |
| HDF5Control | Provides control of aspects of Igor's use of the HDF5 file format. | |
| HDF5Dump | Returns a DDL-format dump of a group, dataset or attribute. | |
| HDF5DumpErrors | Returns information about HDF5-related errors encountered by Igor. This is a diagnostic tool for experts that is needed only in rare cases. | |
HDF5 Procedure Files
Igor ships with two procedure files to support HDF5 use and programming. Both files are automatically loaded by Igor on launch and consequently are always available.
"HDF5 Browser.ipf" implements the HDF5 Browser. This procedure file is an independent module and consequently is normally hidden. If you are an Igor programmer who wants to inspect the procedure file, see Independent Modules for background information. However, there is no reason for you to call routines in "HDF5 Browser.ipf" from your own code.
"HDF5 Utilities.ipf" is a public procedure file (i.e., not an independent module) that defines HDF5-related constants and provides HDF5-related utility routines that may be of use if you write procedures that use HDF5 features.
If you write your own procedure file, you can use the constants and utility routines in "HDF5 Utilities.ipf" without #including anything. However, if you are creating your own independent module for HDF5 programming, you will need to #include "HDF5 Utilities.ipf" into your independent module - see "HDF5 Browser.ipf" for an example.
HDF5 Attributes
An attribute is a piece of data attached to an HDF5 group, dataset or named datatype.
To load an attribute, you need to use the HDF5LoadData operation with the /A=attributeNameStr flag and the /TYPE=objectType flag.
Loading attributes of type H5T_COMPOUND (compound - i.e., structure) is not supported.
Loading an HDF5 Numeric Attribute
This function illustrates how to load a numeric attribute of a group or dataset. The function result is an error code. The value of the attribute is returned via the pass-by-reference attributeValue numeric parameter.
Function LoadHDF5NumericAttribute(pathName, filePath, groupPath, objectName, objectType, attributeName, attributeValue)
String pathName // Symbolic path name - can be "" if filePath is a full path
String filePath // file name or partial path relative to symbolic path, or full path to file
String groupPath // Path to group, such as "/", "/my_group"
String objectName // Name of group or dataset
Variable objectType // 1=group, 2=dataset
String attributeName // Name of attribute
Variable& attributeValue // Output - pass-by-reference parameter
attributeValue = NaN
Variable result = 0
// Open the HDF5 file
Variable fileID // HDF5 file ID will be stored here
HDF5OpenFile /P=$pathName /R /Z fileID as filePath
if (V_flag != 0)
Print "HDF5OpenFile failed"
return -1
endif
Variable groupID // HDF5 group ID will be stored here
HDF5OpenGroup /Z fileID, groupPath, groupID
if (V_flag != 0)
Print "HDF5OpenGroup failed"
HDF5CloseFile fileID
return -1
endif
HDF5LoadData /O /A=attributeName /TYPE=(objectType) /N=tempAttributeWave /Q /Z groupID, objectName
result = V_flag // 0 if OK or non-zero error code
if (result == 0)
Wave tempAttributeWave
if (WaveType(tempAttributeWave) == 0)
attributeValue = NaN // Attribute is string, not numeric
result = -1
else
attributeValue = tempAttributeWave[0]
endif
KillWaves/Z tempAttributeWave
endif
// Close the HDF5 group
HDF5CloseGroup groupID
// Close the HDF5 file
HDF5CloseFile fileID
return result
End
Loading an HDF5 String Attribute
This function illustrates how to load a string attribute of a group or dataset. The function result is an error code. The value of the attribute is returned via the pass-by-reference attributeValue string parameter.
Function LoadHDF5StringAttribute(pathName, filePath, groupPath, objectName, objectType, attributeName, attributeValue)
String pathName // Symbolic path name - can be "" if filePath is a full path
String filePath // file name or partial path relative to symbolic path, or full path to file
String groupPath // Path to group, such as "/", "/metadata_group"
String objectName // Name of group or dataset
Variable objectType // 1=group, 2=dataset
String attributeName // Name of attribute
String& attributeValue // Output - pass-by-reference parameter
attributeValue = ""
Variable result = 0
// Open the HDF5 file
Variable fileID // HDF5 file ID will be stored here
HDF5OpenFile /P=$pathName /R /Z fileID as filePath
if (V_flag != 0)
Print "HDF5OpenFile failed"
return -1
endif
Variable groupID // HDF5 group ID will be stored here
HDF5OpenGroup /Z fileID, groupPath, groupID
if (V_flag != 0)
Print "HDF5OpenGroup failed"
HDF5CloseFile fileID
return -1
endif
HDF5LoadData /O /A=attributeName /TYPE=(objectType) /N=tempAttributeWave /Q /Z groupID, objectName
result = V_flag // 0 if OK or non-zero error code
if (result == 0)
Wave/T tempAttributeWave
if (WaveType(tempAttributeWave) != 0)
attributeValue = "" // Attribute is numeric, not string
result = -1
else
attributeValue = tempAttributeWave[0]
endif
KillWaves/Z tempAttributeWave
endif
// Close the HDF5 group
HDF5CloseGroup groupID
// Close the HDF5 file
HDF5CloseFile fileID
return result
End
Loading All Attributes of an HDF5 Group or Dataset
This function illustrates loading all of the attributes of a given group or dataset. The attributes are loaded into waves in the current data folder.
Function LoadHDF5Attributes(pathName, filePath, groupPath, objectName, objectType, verbose)
String pathName // Symbolic path name - can be "" if filePath is a full path
String filePath // file name or partial path relative to symbolic path, or full path to file
String groupPath // Path to group, such as "/", "/metadata_group"
String objectName // Name of object whose attributes you want or "." for the group specified by groupPath
Variable objectType // The type of object referenced by objectPath: 1=group, 2=dataset
Variable verbose // Bit 0: Print errors; Bit 1: Print warnings; Bit 2: Print routine info
Variable printErrors = verbose & 1
Variable printWarnings = verbose & 2
Variable printRoutine = verbose & 4
Variable result = 0 // 0 means no error
// Open the HDF5 file
Variable fileID // HDF5 file ID will be stored here
HDF5OpenFile /P=$pathName /R /Z fileID as filePath
if (V_flag != 0)
if (printErrors)
Print "HDF5OpenFile failed"
endif
return -1
endif
Variable groupID // HDF5 group ID will be stored here
HDF5OpenGroup /Z fileID, groupPath, groupID
if (V_flag != 0)
if (printErrors)
Print "HDF5OpenGroup failed"
endif
HDF5CloseFile fileID
return -1
endif
HDF5ListAttributes /TYPE=(objectType) groupID, objectName
if (V_Flag != 0)
if (printErrors)
Print "HDF5ListAttributes failed"
endif
HDF5CloseGroup groupID
HDF5CloseFile fileID
return -1
endif
Variable numAttributes = ItemsInList(S_HDF5ListAttributes)
Variable i
for(i=0; i<numAttributes; i+=1)
String attributeNameStr = StringFromList(i, S_HDF5ListAttributes)
STRUCT HDF5DataInfo di
InitHDF5DataInfo(di) // Initialize structure
HDF5AttributeInfo(groupID, objectName, objectType, attributeNameStr, 0, di)
Variable doLoad = 0
switch(di.datatype_class)
case H5T_INTEGER:
case H5T_FLOAT:
case H5T_TIME: // Not yet tested
case H5T_STRING:
case H5T_BITFIELD: // Not yet tested
case H5T_OPAQUE: // Not yet tested
case H5T_REFERENCE:
case H5T_ENUM: // Not yet tested
case H5T_ARRAY: // Not yet tested
doLoad = 1
break
case H5T_COMPOUND: // HDF5LoadData cannot load a compound attribute
doLoad = 0
break
endswitch
if (!doLoad)
if (printWarnings)
Printf "Not loading attribute %s - class %s not supported\r", attributeNameStr, di.datatype_class_str
endif
continue
endif
HDF5LoadData /O /A=attributeNameStr /TYPE=(objectType) /Q /Z groupID, objectName
if (V_flag != 0)
if (printErrors)
Print "HDF5LoadData failed"
endif
result = -1
break
endif
if (printRoutine)
Printf "Loaded attribute %d, name=%s\r", i, attributeNameStr
endif
endfor
// Close the HDF5 group
HDF5CloseGroup groupID
// Close the HDF5 file
HDF5CloseFile fileID
return result
End
HDF5 Dataset Subsets
It is possible, although usually not necessary, to load a subset of an HDF5 dataset using a "hyperslab". To use this feature, you must have a good understanding of hyperslabs which are explained in the HDF5 documentation. The examples below assume that you have read and understood that documentation.
HDF5 does not support loading a subset of an attribute.
To load a subset of a dataset, use the /SLAB flag of the HDF5LoadData operation. The /SLAB flag takes as its parameter a "slab wave". This is a two-dimensional wave containing exactly four columns and at least as many rows as there are dimensions in the dataset you are loading.
The four columns of the slab wave correspond to the start, stride, count and block parameters to the HDF5 H5Sselect_hyperslab routine from the HDF5 library.
The following examples illustrate how to use a hyperslab. The examples use a sample file provided by NCSA and stored in Igor's HDF5 Samples directory.
Create an Igor symbolic path (Misc→New Symbolic Path) named HDF5Samples which points to the folder containing the i32le.h5 file (Igor Pro X Folder:Examples:Feature Demos:HDF5 Samples).
In the first example, we load a 2D dataset named "TestArray" which has 6 rows and 5 columns. We start by loading the entire dataset without using a hyperslab. You can execute the following commands by selecting them and pressing Ctrl-Enter.
Variable fileID
HDF5OpenFile/P=HDF5Samples /R fileID as "i32le.h5"
HDF5LoadData /N=TestWave fileID, "TestArray"
Edit TestWave
Now we create a slab wave and set its dimension labels to make it easier to remember which column holds which type of information. We will use a utility routine in the automatically-loaded "HDF5 Utiliies.ipf" procedure file which is automatically loaded when Igor is launched:
HDF5MakeHyperslabWave("root:slab", 2) // In HDF5 Utilities.ipf
Edit root:slab.ld
Now we set the values of the slab wave to give the same result as before, that is, to load the entire dataset, and then we load the data again using the slab.
slab[][%Start] = 0 // Start at zero for all dimensions
slab[][%Stride] = 1 // Use a stride of 1 for all dimensions
slab[][%Count] = 1 // Use a count of 1 for all dimensions
slab[0][%Block] = 6 // Set block size for the dimension 0 to 6
slab[1][%Block] = 5 // Set block size for the dimension 1 to 5
TestWave = -1 // So we can see the next command change it
HDF5LoadData /N=TestWave /O /SLAB=slab fileID, "TestArray"
Here we set the stride, count and block parameters to load every other element from both dimensions.
slab[][%Start] = 0 // Start at zero for all dimensions
slab[][%Stride] = 2 // Use a stride of 2 for all dimensions
slab[0][%Count] = 3 // Load three blocks of dimension 0
slab[1][%Count] = 2 // Load two blocks of dimension 1
slab[0][%Block] = 1 // Set block size for dimension 0 to 1
slab[1][%Block] = 1 // Set block size for dimension 1 to 1
TestWave = -1 // So we can see the next command change it
HDF5LoadData /N=TestWave /O /SLAB=slab fileID, "TestArray"
Finally we close the file.
HDF5CloseFile fileID
Each row of the slab wave holds a set of parameters (start, stride, count and block) for the corresponding dimension of the dataset in the file. Row 0 of the slab wave holds the parameters for dimension 0 of the dataset, and so on.
The start values must be greater than or equal to zero. All of the other values must be greater than or equal to 1. All values must be less than 2 billion.
HDF5LoadData clips the values supplied for the block sizes to the corresponding sizes of dataset being loaded.
HDF5LoadData requires that the slab wave have exactly four columns. The HDF5MakeHyperslabWave function, from automatically-loaded "HDF5 Utiliies.ipf" procedure file, creates a four-column wave with the column dimension labels Start, Stride, Count, and Block. HDF5LoadData does not require the column dimension labels. As of Igor Pro 9.00, HDF5MakeHyperslabWave returns a free wave if you pass "" for the path parameter. It also returns a wave reference as the function result whether the wave is free or not.
The slab wave must have at least as many rows as the dataset has dimensions. Extra rows are ignored.
HDF5LoadData creates a wave just big enough to hold all of the loaded data. So in the first example, it created a 6 x 5 wave whereas in the last example it created a 3 x 2 wave.
Igor Versus HDF5
This section documents differences in how Igor and HDF5 organize data and how Igor reconciles them.
Case Sensitivity
Igor is not case-sensitive but HDF5 is. So, for example, when you specify the name of an HDF5 data set, case matters. "/Dataset1" is not the same as "/dataset1".
If you load /Dataset1 and /dataset1 into Igor using the default wave name, the second load overwrites the wave created by the first.
HDF5 Object Names Versus Igor Object Names
The forward slash character is not allowed in HDF5 object names. If you create an Igor wave or data folder with a name containing a forward slash and attempt to save the object to an HDF5 file, you will get an error. An object name that starts with a dot may also create an error.
As of this writing, the section 4.2.3, "HDF5 Path Names" of the HDF5 User's Guide says:
Component link names may be any string of ASCII characters not containing a slash or a dot (/and ., which are reserved as noted above). However, users are advised to avoid the use of punctuation and non-printing characters, as they can create problems for other software.
HDF5 Data Types Versus Igor Data Types
HDF5 supports many data types that Igor does not support. When loading such data types, Igor attempts to convert to a data type that it supports, if possible. In the process, precision may be lost.
By default and for backward compatibility, Igor saves and loads HDF5 64-bit integer data as double-precision floating point. Precision may be lost in this conversion. To save and load 64-bit integer data as 64-bit integer, use /OPTS=1 with HDF5SaveData, HDF5SaveGroup, HDF5LoadData and HDF5LoadGroup. Because most operations are carried out in Igor in double-precision floating point, we recommend loading 64-bit integer as double if it fits in 53 bits and as 64-bit integer if it may exceed 53 bits.
Since Igor does not currently support 128-bit floating point data (long double), Igor loads HDF5 128-bit floating point data as double-precision floating point. Precision is lost in this conversion.
HDF5 Max Dimensions Versus Igor Max Dimensions
The HDF5 library supports data with up to 32 dimensions. Igor supports only four dimensions. If you load HDF5 data whose dimensionality is greater than four, the HDF5LoadData operation creates a 4D wave with enough chunks to hold all of the data. ("Chunk" is the name of the fourth dimension in Igor, after "row", "column" and "layer".)
For example, if you load a dataset whose dimensions are 7x6x5x4x3x2, HDF5LoadData creates a wave with dimensions 7x6x5x24. The wave has 24 chunks. The number 24 comes from multiplying the number of chunks in the HDF5 dataset (4 in this case) by the size of each higher dimension (3 and 2 in this case).
Row-major Versus Column-major Data Order
HDF5 and Igor store multi-dimensional data differently in memory. For almost all purposes, this difference is immaterial. For those rare cases in which it matters, here is an explanation.
HDF5 stores multi-dimensional data in "row-major" order. This means that the index associated with the highest dimension changes fastest as you move in memory from one element to the next. For example, if you have a two-dimensional dataset with 5 rows and 4 columns, as you move sequentially through memory, the column index changes fastest and the row index slowest.
Igor stores data in "column-major" order. This means that the index associated with the lowest dimension changes fastest as you move in memory from one element to the next. For example, if you have a two-dimensional dataset with 5 rows and 4 columns, as you move sequentially through memory, the row index changes fastest and the column index slowest.
To work around this difference, after the HDF5LoadData or HDF5LoadGroup operation initially loads data with two or more dimensions, it shuffles the data around in memory. The result is that the data when viewed in an Igor table looks just as it would if viewed in the HDF Group's HDFView program although its layout in memory is different. A similar accomodation occurs when you save Igor data using HDF5SaveData or HDF5SaveGroup.
HDF5 Images Versus Igor Images
The HDF5 Image and Palette Specification provides detailed guidelines for storing an image and its associated information (e.g., palette, color model) in an HDF5 file. However many HDF5 users do not follow the specification and just write image data to a regular 2D dataset. To distinguish between these two ways of storing an image, we use the term "formal image" to refer to an image written to the specification and "regular image" to refer to regular 2D datasets which the user thinks of as an image.
In an Igor image plot the wave's column data is plotted horizontally while in HDFView and most other programs the row data is plotted horizontally. Therefore, without special handling, a regular image would appear rotated in Igor relative to most programs.
The HDF5LoadImage and HDF5SaveImage operations handle loading and saving formal images. These operations automatically compensate for the difference in image orientation.
If you are dealing with a regular image, you will use the HDF5LoadData and HDF5SaveData operations, or HDF5LoadGroup and HDF5SaveGroup. These operations have a /TRAN flag which causes 2D data to be transposed. When you use /TRAN with HDF5LoadData, images viewed in Igor and in programs like HDFView will have the same orientation but will appear transposed when viewed in a table.
The /TRAN flag works with 2D and higher-dimensioned data. When used with higher-dimensioned data (3D or 4D), each layer of the data is treated as a separate image and is transposed. In other words, /TRAN treats higher-dimensioned data as a stack of images.
Saving and Reloading Igor Data
The HDF5SaveData and HDF5SaveGroup operations can save Igor waves, numeric variables and string variables in HDF5 files. All of these Igor objects are written as HDF5 datasets.
The datasets saved from Igor waves are, by default, marked with attributes that store wave properties such as the wave data type, the wave scaling and the wave note. The attributes have names like IGORWaveType and IGORWaveScaling. This allows HDF5LoadData and HDF5LoadGroup to fully recreate the Igor wave if it is later read from the HDF5 file back into Igor. You can suppress the creation of these attributes by using the /IGOR=0 flag when calling HDF5SaveData or HDF5SaveGroup.
Wave text is always written using UTF-8 text encoding. See HDF5 Wave Text Encoding for details.
Wave reference waves and data folder reference waves are read as such when you load an HDF5 packed experiment but HDF5LoadData and HDF5LoadGroup load these waves as double-precision numeric. The reason for this is that restoring such waves so that they point to the correct wave or data folder is is possible only when an entire experiment is loaded.
The datasets saved by HDF5SaveGroup from Igor variables are marked with an "IGORVariable" attribute. This allows HDF5LoadData and HDF5LoadGroup to recognize these datasets as representing Igor variables if you reload the file. In the absence of this attribute, these operations load all datasets as waves.
The value of the IGORVariable attribute is the data type code for the Igor variable. It is one of the following values:
| 0: | Igor string variable | |
| 4: | Igor real numeric variable | |
| 5: | Igor complex numeric variable | |
See also HDF5 String Variable Text Encoding.
Handling of Complex Waves
Igor Pro supports complex waves but HDF5 does not support complex datasets. Therefore, when saving a complex wave, HDF5SaveData writes the wave as if its number of rows were doubled. For example, HDF5SaveData writes the same data to the HDF5 file for these waves:
Make wave0 = {1,-1,2,-2,3,-3} // 6 scalar points
Make/C cwave0 = {cmplx(1,-1),cmplx(2,-2),cmplx(3,-3)} // 3 complex points
When reading an HDF5 file written by HDF5SaveData, you can determine if the original wave was complex by checking for the presence of the IGORComplexWave attributes that HDF5SaveData attaches to the dataset for a complex wave. HDF5LoadData and HDF5LoadGroup do this automatically if you use the appropriate /IGOR flag.
Handling of Igor Reference Waves
Igor Pro supports wave reference waves and data folder reference waves. Each element of a wave reference wave is a reference to another wave. Each element of a data folder reference wave is a reference to a data folder.
Igor correctly writes Igor reference waves when you save an experiment as HDF5 packed format, but the HDF5SaveData and HDF5SaveGroup do not support saving Igor reference waves.
The HDF5SaveData operation returns an error if you try to save a reference wave.
The behavior of the HDF5SaveGroup operation when it is asked to save a reference wave depends on the /CONT flag. By default (/CONT=1), it prints a note in the history saying it cannot save the wave and then continues saving the rest of the objects in the data folder. If /CONT=0 is used, HDF5SaveGroup returns an error if asked to save a reference wave.
HDF5 Multitasking
You can call HDF5 operations and functions from an Igor preemptive thread.
The HDF5 library is limited to accessing one HDF5 file at a time. This precludes loading multiple HDF5 files concurrently in a given Igor instance but it does allow you to load an HDF5 file in a preemptive thread while you do something else in Igor's main thread. If you create multiple threads that attempt to access HDF5 files, one of your threads will gain access to the HDF5 library. Your other HDF5-accessing threads will wait until the first thread finishes at which time another thread will gain access to the HDF5 library.
Igor HDF5 Capabilities
Igor supports only a subset of all of the HDF5 capability.
Here is a partial list of HDF5 features that Igor does support:
- Loading of all atomic datatypes.
- Loading of strings.
- Loading of array datatypes.
- Loading of variable-length datasets where the base type is integer or float.
- Loading of compound datasets (datasets consisting of C-like structures), including compound datasets containing members that are arrays.
- Use of hyperslabs to load subsets of datasets.
- Loading and saving object references.
- Loading dataset region references.
Igor HDF5 Limitations
Here is a partial list of HDF5 features that Igor does not support:
- Creating or appending to VLEN datasets (ragged arrays).
- Loading of deeply-nested datatypes. See HDF5 Nested Datatypes below.
- Saving dataset region references.
If Igor does not work with your HDF5 file, it could be due to a limitation in Igor. Send a sample file along with a description of what you are trying to do to support@wavemetrics.com and we will try to determine what the problem is.
Advanced HDF5 Data Types
This section is mostly of interest to advanced HDF5 users.
HDF5 Variable-Length Data
Most HDF5 files do not use variable-length datatypes so most users do not need to know this information.
Variable-length data consists of an array where each element is a 1D set of elements of another datatype, called the "base" datatype. The number of elements of the base datatype in each set can be different. For example, a 5 element variable-length dataset whose base type is H5T_STD_I32LE contains 5 1D sets of 32-bit, little-endian integers and the length of each set is independent.
HDF5LoadData loads variable-length datasets where the base type is integer or float only. The data for each element is loaded into a separate wave.
When loading most types of data, HDF5LoadData creates just one wave. When loading a variable-length dataset or attribute, one wave is created for each loaded element. If more than one element is to be loaded, the proposed name for the wave (the name of the dataset or attribute being loaded or a name specified by /N=name ) is treated as a base name. For example, if the dataset or attribute has three elements and name is test and the /O flag is used, waves named test0, test1 and test2 are created. If the /O flag is not used, names of the form test<n> are created where <n> is a number chosen to make the wave names unique.
HDF5LoadData operation supports loading a subset of a variable-length dataset. You do this by supplying a slab wave using the HDF5LoadData /SLAB flag. In the example from the previous paragraph, if you loaded just one element, its name would be test, not test0. If you loaded two elements, they would be named test0 and test1, regardless of which two elements you loaded.
This function demonstrates loading one element of a variable-length dataset. We assume that a symbolic path named Data and a file named "Vlen.h5" exist and that the file contains a 1D variable-length dataset named TestVlen that contains at least two elements. The function loads the second variable-length element into a wave named TestWave.
Function DemoVlenLoad()
Variable fileID
HDF5OpenFile /P=Data /R fileID as "Vlen.h5"
HDF5MakeHyperslabWave("root:slab", 1) // In HDF5 Utilities.ipf
Wave slab = root:slab
slab[0][%Start] = 1 // Start at second vlen element
slab[0][%Stride] = 1 // Use a stride of 1
slab[0][%Count] = 1 // Load 1 block
slab[0][%Block] = 1 // Set block size to 1
HDF5LoadData /N=TestWave /O /SLAB=slab fileID, "TestVlen"
HDF5CloseFile fileID
End
HDF5 Array Data
Most HDF5 files do not use array datatypes so most users do not need to know this information.
An HDF5 dataset (or attribute) consists of elements organized in one or more dimensions. Each element can be an atomic datatype, such as an unsigned short or a double-precision float, or it can be a composite datatype, such as a structure or an array. Thus, an HDF5 dataset can be an array of unsigned shorts, an array of doubles, an array of structures or an array of arrays. This section discusses loading this last type - an array of arrays.
In this case, the class of the dataset is H5T_ARRAY. The type of the dataset is something like "5 x 4 array of signed long" and "signed long" is said to be the "base type" of the array datatype. If the dataset itself is 1D with 10 rows then you would have a 10-row dataset, each element of which consists of one 5 x 4 array of signed longs.
In Igor this could be treated as a 3D wave consisting of 5 rows, 4 columns and 10 layers. However, since Igor supports only 4 dimensions while HDF5 supports 32, you could easily run out of dimensions in Igor. Therefore when you load an H5T_ARRAY-type dataset, HDF5LoadData creates a wave whose dimensionality is that of the array type, not that of the underlying dataset, except that the highest dimension is increased to make room for all of the data. This reduces the likelihood of running out of dimensions.
HDF5LoadData cannot load array data whose base type is compound or array (see HDF5 Nested Datatypes for details).
The following example illustrate how to load an array datatype. The example uses a sample file provided by NCSA and stored in Igor's HDF5 Samples directory.
Create an Igor symbolic path (Misc→New Symbolic Path) named HDF5Samples which points to the folder containing the SDS_array_type.h5 file (Igor Pro X Folder:Examples:Feature Demos:HDF5 Samples).
The sample file contains a 10 row 1D data set, each element of which is a 5 row by 4 column matrix of 32-bit signed big-endian integers (a.k.a., big-endian signed longs or I32BE).
When we load the dataset, we get 5 rows and 40 columns.
Variable fileID
HDF5OpenFile/P=HDF5Samples /R fileID as "SDS_array_type.h5"
HDF5LoadData fileID, "IntArray"
HDF5CloseFile fileID
Edit IntArray
The first four columns contain the 5x4 matrix from row zero of the dataset. The next four columns contain the 5x4 matrix from row one of the dataset. And so on.
In this case, Igor has enough dimensions so that you could, if you want, reorganize it as a 3D wave consisting of 10 layers of 5x4 matrices. You would do that using this command:
Redimension /N=(5,4,10) /E=1 IntArray
The /E=1 flag tells Redimension to change the dimensionality of the wave without actually changing the stored data. In Igor, the layout in memory of a 5x4x10 wave is the same as the layout of a 5x40 wave. The redimension merely changes the way we look at the wave from 40 columns of 5 rows to 10 layers of 4 columns of 5 rows.
Although you can load a dataset with an array datatype, Igor currently provides no way to write a datatype with an array datatype.
Loading HDF5 Reference Data
Most HDF5 files do not use reference datatypes so most users do not need to know this information.
An HDF5 dataset or attribute can contain references to other datasets, groups and named datatypes. There are two types of references: "object references" and "dataset region references". HDF5LoadData loads both types of references into text waves.
An element of an object reference dataset can refer to a dataset in the same or in another file. An element of a dataset region reference dataset can refer only to a dataset in the same file.
Loading HDF5 Object Reference Data
For each object reference to a dataset, HDF5LoadData returns "D:" plus the full path of the dataset within the HDF5 file, for example, "D:/GroupA/DatasetB".
For references to groups and named datatypes, HDFLoadData returns "G:" and "T:" respectively, followed by the path to the object.
Loading HDF5 Dataset Region Reference Data
This section is for advanced HDF5 users. A demo experiment, "HDF5 Dataset Region Demo.pxp", provides examples and utilities for dealing with dataset region references.
Prior to Igor Pro 9.00, HDF5LoadData returned the same thing when loading an object reference or a dataset region reference. It had this form:
<object type character>:<full path>
For a dataset, this might be something like
D:/Group1/Dataset3
In Igor Pro 9.00 and later, HDF5LoadData returns additional information for a dataset region reference. It has this form with the additional information shown in red:
<object type character>:<full path>.<region info>
For a dataset, this might be something like this with the additional information shown in red:
D:/Group1/Dataset3.REGIONTYPE=BLOCK;NUMDIMS=2;NUMELEMENTS=2;COORDS=0,0-0,2/0,11-0,13;
If you have code that depends on the pre-Igor 9 behavior, you can make it work with Igor 9 by using StringFromList with "." as list separator string to obtain the text preceding the dot character.
The region info string is a semicolon-separated keyword-value string constructed for ease of programmatic parsing. It consists of these parts:
REGIONTYPE=<type>
NUMDIMS=<number of dimensions>
NUMELEMENTS=<number of elements>
COORDS=<list of points> or <list of blocks>
<type> is POINT for a region defined as a set of points, BLOCK for a region defined as a set of blocks.
<number of dimensions> is the number of dimensions in the dataset.
<number of elements> is the number of points in a region defined as a set of points or the number of blocks in a region defined as a set of blocks.
<list of points> has the following format:
<point coordinates>/<point coordinates>/...
where <point coordinates> is a comma-separated list of coordinates.
For a 2D dataset with three selected points, this might look like this:
3,4/11,13/21,30
which specifies these three points:
row 3, column 4
row 11, column 13
row 21, column 30
<list of blocks> has the following format:
<coordinates>-<coordinates>/<coordinates>-<coordinates>/...
where <coordinates> is a comma-separated list of coordinates.
A dash appears between pairs of <coordinates>. The first set of coordinates of a pair specifies the starting coordinates of a block while the second set of coordinates of a pair specifies the ending coordinates of the block.
For a 2D dataset with three selected blocks, this might look like this:
3,4-6,7/11,13-15,17/21,30-37,38
which specifies these three blocks:
row 3, column 4 to row 6, column 7
row 11, column 13 to row 15, column 17
row 21, column 30 to row 37, column 38
Here is an example of a complete point dataset region info string with the additional information shown in red:
D:/Group1/Dataset3.REGIONTYPE=POINT;NUMDIMS=2;NUMELEMENTS=3;COORDS=3,4/11,13/21,30;
Here is an example of a complete block dataset region info string with the additional information shown in red:
D:/Group1/Dataset4.REGIONTYPE=BLOCK;NUMDIMS=2;NUMELEMENTS=3;COORDS=3,4-6,7/11,13-15/17/21,30-37/38;
The wave returned after calling HDF5LoadData on a two-row dataset region reference dataset would contain two rows of text like the examples just shown. Each row in the dataset region reference dataset refers to one dataset and to a set of points or blocks within that dataset.
The "HDF5 Dataset Region Demo.pxp" experiment provides further information including examples and utilities for dealing with dataset region references.
Saving HDF5 Object Reference Data
Most HDF5 files do not use reference datatypes so most users do not need to know this information.
An HDF5 dataset or attribute can contain references to other datasets, groups and named datatypes. These are called "object references". You can instruct HDF5SaveData to save a text wave as an object reference using the /REF flag. The /REF flag requires Igor Pro 8.03 or later.
The text to save as a reference must be formatted with a prefix character identifying the type of the referenced object followed by a full or partial path to the object: "G:", "D", or "T:" for groups, datatypes, and datasets respectively. For example:
Function DemoSaveReferences(pathName, fileName)
String pathName // Name of symbolic path
String fileName // Name of HDF5 file
Variable fileID
HDF5CreateFile/P=$pathName /O fileID as fileName
// Create a group to target using a reference
Variable groupID
HDF5CreateGroup fileID, "GroupA", groupID
// Create a dataset to target using a reference
Make/O/T textWave0 = {"One", "Two", "Three"}
HDF5SaveData /O /REF=(0) /IGOR=0 textWave0, groupID
// Write reference dataset to root using full paths
Make/O/T refWaveFull = {"G:/GroupA", "D:/GroupA/textWave0"}
HDF5SaveData /O /REF=(1) /IGOR=0 refWaveFull, fileID
HDF5CloseGroup groupID
HDF5CloseFile fileID
End
Partial paths are relative to the file ID or group ID passed to HDF5SaveData.
Saving HDF5 Dataset Region Reference Data
Igor Pro does not currently support saving dataset region references.
Loading HDF5 Enum Data
Most HDF5 files do not use enum datatypes so most users do not need to know this information.
Enum data values are stored in an HDF5 file as integers. The datatype associated with an enum dataset or attribute defines a mapping from an integer value to a name. HDF5LoadData can load either the integer data or the name for each element of enum data. You control this using the /ENUM=enumMode flag.
If /ENUM is omitted or enumMode is zero, HDF5LoadData creates a numeric wave with data type signed long and loads the integer enum values into it. This works for 8 bit, 16 bit and 32 bit integer enum data. Loading enum data based on 64-bit integers is not supported.
If enumMode is 1, HDF5LoadData creates a text wave and loads the name associated with each enum value into it. This is slower than loading the integer enum values but the speed penalty is significant only if you are loading a very large enum dataset or very many enum datasets.
Saving HDF5 Enum Data
Most HDF5 files do not use enum datatypes so most users do not need to know this information.
The ability to save Igor integer numeric data using an HDF5 enumeration data type was added in Igor Pro 9.01. You do this using the /ENUM flag of the HDF5SaveData operation. Neither the HDF5 Browser nor the HDF5SaveGroup support saving enum data.
Here is an example:
Function DemoSaveWaveAsEnum()
Variable fileID
HDF5CreateFile/P=IgorUserFiles /O fileID as "DemoSaveWaveAsEnum.h5"
Make/FREE/Y=(0x48) enumWave = {1,2,3} // 0x48 means unsigned byte data
String enumList = "One=1;Two=2;Three=3;" // Specifies the HDF5 enum datatype
HDF5SaveData /O /ENUM=enumList /IGOR=0 enumWave, fileID, "enumDataset"
HDF5CloseFile fileID
End
This example writes a dataset named "enumDataset" with a base type of unsigned byte (/Y=(0x48)). The enum datatype defines three names, One, Two, and Three, associated with three integer values, 1, 2 and 3.
The syntax for enumList is very strict. For each enumeration datatype member, you must supply one name followed by an equal sign followed by an integer number followed by a semicolon. No white space or any other characters are permitted.
The wave being saved must have an integer type: 8-bit, 16-bit, 32-bit or 64-bit signed or unsigned integer. HDF5LoadData cannot load 64-bit integer enum data so don't use 64 bits if you need to load the data back into Igor.
HDF5 Opaque Data
Most HDF5 files do not use opaque datatypes so most users do not need to know this information.
Opaque data consists of elements that are treated as a string of bytes of a specified length. HDF5LoadData loads opaque data into an unsigned byte wave. If an element of opaque data is n bytes then each element occupies n contiguous rows in the wave.
HDF5 Bitfield Data
Most HDF5 files do not use bitfield datatypes so most users do not need to know this information.
Bitfield data consists of elements that are a sequence of bits treated as individual values. HDF5LoadData loads bitfield data of any length into an unsigned byte wave with as many bytes per element as are needed. For example, if the bitfield is two bytes then two rows of the unsigned byte wave are used for each element.
The data is loaded using the byte order as stored in the file. This is appropriate if you think of the data as a stream of bytes. If you think of the data as a short (2 bytes) or long (4 bytes), you can use the Redimension command. For example, if you just loaded a 2D data set containing 5 two-byte bitfields per row and 3 columns, you wind up with a 10x3 unsigned byte wave. You can change it to a 5x3 wave of shorts like this:
Redimension /N=(5,3) /W /E=1 bitfieldWave
If you need to change the byte order, use /E=2 instead of /E=1.
HDF5 Nested Datatypes
Most HDF5 files do not use nested datatypes so most users do not need to know the following information.
HDF5 supports "atomic" data classes, such as integers (called class H5T_INTEGER in the HDF5 library) and floats (class H5T_FLOAT), and "composite" data classes, such as structures (class H5T_COMPOUND), arrays (class H5T_ARRAY) and variable-length (class H5T_VLEN) types.
In a dataset whose datatype is atomic, each element of the dataset is a single value. In a dataset whose datatype is composite, each element is a collection of elements of one or more other datatypes.
For example, consider a 5x3 array of data of type H5T_STD_I16BE. H5T_STD_I16BE is a built-in atomic datatype of class H5T_INTEGER. Each element of such an array is a 16-bit, big-endian integer value. This dataset has an atomic datatype.
Now consider a 4 row dataset of class H5T_ARRAY where each element of the dataset is a 5x3 array of data of type H5T_STD_I16BE. This dataset has a composite datatype. Each of the 4 elements of the dataset is an array, in this case, an array of 16-bit, big-endian integer values. H5T_STD_I16BE is the "base datatype" of each of the 4 arrays.
Now consider a 4 row dataset of class H5T_COMPOUND with one integer member, one float member and one string member. This dataset has a composite datatype. Each of the 4 elements of this dataset is a structure with members of three different datatypes.
In HDF5 it is possible to nest datatypes to any depth. For example, you can create an H5T_ARRAY dataset where each element is an H5T_COMPOUND dataset where the members are H5T_ARRAY datasets of H5T_STD_I16BE, H5T_STD_I32BE and other datatypes.
Igor does not support indefinitely nested datasets. It supports only the following:
- Atomic datasets of almost any type.
- Array datasets where the base type is integer, float, string, bitfield, opaque or enum.
- Compound datasets where the member's base type is integer, float, string, bitfield, opaque, enum or reference.
- Compound datasets with array members where the base type of the array is integer, float, string, bitfield, opaque, enum or reference.
- Variable-length datasets where the base type is integer or float.
HDF5 Compound Data
This is an advanced feature that most users will not need.
In an HDF5 compound dataset, each element of the dataset consists of a set of named fields, like an instance of a C structure. Loading compound datasets is problematic because their structure can be arbitrarily complex.
A compound data set may contain a collection of disparate datatypes, arrays of disparate datatypes, and sub-compound structures.
HDF5LoadData can load either a single member from a compound dataset into a single wave, or it can load all members of the compound dataset into a set of waves. However, if a member is too complex, HDF5LoadData cannot load it and returns an error. For the most part, "too complex" means that the member is itself compound (a sub-structure).
You instruct the HDF5LoadData operation to load a single member from each element of the compound dataset by using the /COMP flag with a mode parameter set to one and with the name of the member to load. The member must be an atomic datatype or an array datatype but cannot be another compound datatype (see HDF5 Nested Datatypes for details). HDF5LoadData creates an Igor wave with a data type that is compatible with the datatype of the HDF5 dataset. The name of the wave is based on the name of the dataset or attribute being loaded or on the name specified by the /N flag.
You instruct HDF5LoadData to load all members into separate waves by omitting the /COMP flag or specifying /COMP={0,""}. The names of the waves created consist of a base name concatenated with a possibly cleaned up version of the member name. The base name is based on the name of the dataset or attribute being loaded or on the name specified by the /N flag.
Although you can load an HDF5 dataset with a compound datatype, Igor currently provides no way to write a datatype with a compound datatype.
HDF5 Compression
Igor can use compression when writing datasets to HDF5 files. Compressed datasets usually take less space on disk but more time to write and more time to read.
The amount of disk space saved by compression depends on the size and nature of your data. If your data is very noisy, compression may save little disk space or even increase the amount of disk space required. If your data is nearly all zero or some other constant value, compression may save 99 percent of disk space. With typical real-world data, compression may save from 10 to 50 percent of disk space.
The time required to save a compressed dataset is typically in the range of 2 to 10 times the time required to save an uncompressed dataset. The time required to load a compressed dataset is typically 2 times the time required to load an uncompressed dataset. These times can vary widely depending on your data.
Compression may be performed when you
- Save an HDF5 packed experiment file
- Save a dataset or group using the HDF5 Browser
- Call the HDF5SaveData operation
- Call the HDF5SaveGroup operation
To enable compression Igor must provide certain parameters to the HDF5 library:
-
The level of compression for GZIP from 0 to 9
-
Whether shuffling should be performed or not
-
The chunk size to use for each dimension of the wave being saved
Compression parameters for a given dataset are set when the dataset is created and cannot be changed when appending to an existing dataset.
There are several ways through which Igor obtains these parameters:
-
From the HDF5SaveData operation /GZIP and /LAYO flags
-
From the HDF5SaveGroup, SaveExperiment, and SaveData operation /COMP flags
-
From the SaveGraphCopy, SaveTableCopy, and SaveGizmoCopy operation /COMP flags
-
From the HDF5SaveDataHook (see Using HDF5SaveDataHook for details)
-
Via HDF5 default compression for manual saves (see HDF5 Default Compression for details)
HDF5 supports compression of datasets through three categories of filters:
-
Internal filters: Shuffle, Fletcher32, ScaleOffset, and NBit
Internal filters are implemented in the HDF5 library source code.
-
External filters: GZIP, SZIP
External filters are linked to the HDF5 library when the library is compiled.
-
Third-party filters
Third-party filters are detected and loaded by the HDF5 library at runtime.
Igor Pro currently supports the following HDF5 filters:
-
GZIP
-
SZIP for reading only - not supported for writing
-
Shuffle
Shuffle reorders the bytes of multi-byte data elements and can result in higher compression ratios.
HDF5 Layout Chunk Size
Layout refers to how data for a given dataset is arranged on disk. The HDF5 library supports three types of layout: contiguous, compact, chunked. The HDF5SaveData and HDF5SaveGroup operations default to contiguous layout which is appropriate for uncompressed datasets.
The HDF5 library requires that compressed datasets use the chunked layout. This means that the data for the dataset is written in some number of discrete chunks rather than in one contiguous block. In the context of compression, chunking is mainly useful for speeding up accessing subsets of a dataset. Without chunking, the entire dataset has to be read and decompressed to read any subset. If the dataset is always read in its entirety, such as when Igor loads an HDF5 packed experiment file, chunking does not enhance speed.
Chunked storage requires telling the HDF5 library the size of a chunk for each dimension of the dataset. There is no general way to choose appropriate chunk sizes because it depends on the dimensions and nature of the data as well as tradeoffs that require the judgement of the user. In Igor chunk sizes can be specified
-
Using the HDF5SaveData operation /LAYO flag (see HDF5SaveData for details)
-
Using HDF5SaveDataHook (see Using HDF5SaveDataHook for details)
The HDF5SaveGroup, SaveExperiment, SaveData, SaveGraphCopy, SaveTableCopy, and SaveGizmoCopy operations and HDF5 default compression always save a compressed dataset in one chunk, as described in the following sections.
Compression parameters for a given dataset are set when the dataset is created and cannot be changed when appending to an existing dataset.
HDF5SaveGroup Compression
In Igor Pro 9.00 and later, you can tell the HDF5SaveGroup operation to compress numeric datasets using the /COMP flag.
HDF5SaveGroup applies compression only to numeric waves, not to text waves or other non-numeric waves nor to numeric waves with fewer than the number of elements specified by the /COMP flag.
HDF5SaveGroup compression uses chunk sizes equal to the size of each dimension of the wave. Such chunk sizes mean that the entire wave is written using chunked layout as one chunk. This is fine for datasets that are to be read all at once. For finer control you can use the HDF5SaveDataHook function to override compression specified by /COMP but this is usually not necessary.
SaveExperiment Compression
In Igor Pro 9.00 and later, you can tell the SaveExperiment operation to compress numeric datasets using the /COMP flag. This works the same as HDF5SaveGroup compression discussed in the preceding section.
The same is true for the SaveData, SaveGraphCopy, SaveTableCopy, and SaveGizmoCopy operations.
See HDF5 Compression for Saving Experiment Files for step-by-step instructions.
HDF5 Default Compression
Igor can perform default dataset compression when you save HDF5 files via the user interface. Default compression does not apply when saving HDF5 files via the HDF5SaveData, HDF5SaveGroup, SaveExperiment, SaveData, SaveGraphCopy, SaveTableCopy, or SaveGizmoCopy operations. HDF5 default compression was added in Igor Pro 9.00.
Default compression is disabled by default because compression can significantly increase the time required to save. Whether compression is worthwhile depends on the size and nature of your data and on how you trade off time versus disk space.
You can enable default compression by choosing Misc→Miscellaneous Settings and clicking the Experiment icon. This allows you to set the following settings:
- Whether default compression is enabled or disabled
- The minimum size a wave must be before compression is used
- A compression level from 0 (no compression) to 9 (max)
- Whether you want to enable shuffle (an optional process before compression)
You can programmatically determine the default compression currently in effect like this:
Print IgorInfo(14) // Print default compression settings
Once you have turned default compression on, it applies to saving HDF5 packed experiment files via the user interface, namely via the Data Browser Save Copy button, via the HDF5 Browser Save Waves and Save Data Folder buttons, and via the following File menu items: Save Experiment, Save Experiment As, Save Experiment Copy, Save Graph Copy, Save Table Copy, and Save Gizmo Copy.
The HDF5SaveDataHook function, if it exists, can override default compression settings.
Igor applies default compression only to numeric waves, not to text waves or other non-numeric waves nor to numeric waves with fewer than the number of elements specified by the settings.
Default compression uses chunk sizes equal to the size of each dimension of the wave. Such chunk sizes mean that the entire wave is written using chunked layout as one chunk. This is fine for datasets that are to be read all at once.
Igor's HDF5 default compression is sufficient for most purposes. If necessary, for example if you intend to read subsets rather than the entire dataset at once, you can override it using the HDF5SaveData operation or an HDF5SaveDataHook function.
Using HDF5SaveDataHook
HDF5SaveDataHook is a user-defined function that Igor calls to determine what kind of compression, if any, should be applied when saving a given wave as a dataset.
Support for HDF5SaveDataHook was added in Igor Pro 9.00.
HDF5SaveDataHook is an experimental feature for advanced users only. The feature may be changed or removed. If you find it useful, please let us know, and send your function and an explanation of what purpose it serves.
The hook function takes a single HDF5SaveDataHookStruct parameter containing input and output fields. The version, operation, and waveToSave fields are inputs. The gzipLevel, shuffle, and chunkSizes fields are outputs.
The main use for HDF5SaveDataHook is to provide a way by which you can specify chunk sizes on a wave-by-wave basis taking into account the wave dimension sizes and your tradeoff of disk space versus save time. The HDF5SaveData operation allows you to specify chunk sizes via the /LAYO flag. However, the other operations that support HDF5 compression through the /COMP flag (HDF5SaveGroup, SaveExperiment, SaveData, SaveGraphCopy, SaveTableCopy, and SaveGizmoCopy) always save datasets as one chunk as does HDF5 default compression. HDF5SaveDataHook allows you to change that, though in most cases there is no need to.
If the operation field is non-zero, then the hook function is being called from the HDF5SaveData or HDF5SaveGroup operations. You should respect the compression settings specified to those operations unless there is good reason to override them.
To set the compression parameters for the specified wave, set the output fields and return 1. To allow the save to proceed without altering compression, return 0.
Here is an example:
ThreadSafe Function HDF5SaveDataHook(s)
STRUCT HDF5SaveDataHookStruct& s
// Print "Global ThreadSafe HDF5SaveDataHook called" // For debugging only
// return 0 // Uncomment this to make this hook a NOP
if (s.version >= 2000) // Structure version is 1000 as of this writing
return 0 // The structure changed in an incompatible way
endif
if (s.operation != 0) // Called from HDF5SaveData or HDF5SaveGroup operations?
return 0 // Respect their settings unless we have good reason to change them
endif
// Set compression parameters only for numeric waves
int isNumeric = WaveType(s.waveToSave) == 1
if (!isNumeric)
return 0
endif
// Set compression parameters only if the total number of elements
// in the wave exceeds a particular threshold
int numElements = numpnts(s.waveToSave)
if (numElements < 10000)
return 0
endif
// Set compression parameters
s.gzipLevel = 5 // 0 (no compression) to 9 (max compression)
s.shuffle = 0 // 0=shuffle off, 1=shuffle on
s.chunkSizes[0] = 200 // These values are arbitrary. Igor clips them.
s.chunkSizes[1] = 200 // In a real function you would have some more
s.chunkSizes[2] = 50 // systematic way to choose them.
s.chunkSizes[3] = 50
// Indicate that we want to set compression parameters for this wave
return 1
End
The HDF5SaveDataHook function must be threadsafe. If it is not threadsafe, Igor will not call it.
The HDF5SaveDataHook function is called when a dataset is created and not when appending to an existing dataset.
The HDF5SaveDataHook function must not alter the state of Igor. That is, it must have no side effects such as creating waves or variables, modifying waves or variables, or killing waves or variables. It is called when HDF5SaveData is called, when HDF5SaveGroup is called, and also when Igor is saving an HDF5 packed experiment file. If you change the state of Igor in your HDF5SaveDataHook function, this may cause file corruption or a crash.
It is possible but not recommended to define more than one HDF5SaveDataHook function. For example, you can define one in the ProcGlobal context, another in a regular module and a third in an independent module. If you do this, each function is called until one returns 1. The order in which the functions are called is not defined.
We recommend that you define your HDF5SaveDataHook function in an independent module so that it can be called when normal procedures are in an uncompiled state. Otherwise, if you save an HDF5 packed experiment while procedures are not compiled, for example because of an error, your HDF5SaveDataHook function will not be called and all datasets will be saved with default compression or uncompressed.
For testing purposes, you can prevent Igor from calling any HDF5SaveDataHook function by executing:
SetIgorOption HDF5SaveDataHook=0
This prevents Igor from calling the hook function via HDF5SaveData and HDF5SaveGroup operations and when saving an HDF5 packed experiment file.
You can re-enable calling the hook function by executing:
SetIgorOption HDF5SaveDataHook=1
HDF5 Compression References
This section lists documents that discuss HDF5 filtering and compression that may be of interest to advanced HDF5 users.
-
Using Compression in HDF5
https://support.hdfgroup.org/documentation/hdf5-docs/hdf5_topics/UsingCompressionInHDF5.html
-
Chunking in HDF5
https://support.hdfgroup.org/documentation/hdf5-docs/advanced_topics/chunking_in_hdf5.html
-
HDF5 Advanced Topics:Chunking in HDF5
-
Dataset Chunking Issues
https://support.hdfgroup.org/documentation/hdf5/latest/hdf5_chunk_issues.html
-
HDF5 Compression Demystified #1
https://www.hdfgroup.org/2015/04/hdf5-data-compression-demystified-1/
-
HDF5 Compression Demystified #2
https://www.hdfgroup.org/2017/05/hdf5-data-compression-demystified-2-performance-tuning/
-
Improving I/O Performance When Working with HDF5 Compressed Datasets
-
HDF5 Compression Troubleshooting
https://support.hdfgroup.org/documentation/hdf5-docs/hdf5_topics/HDF5CompressionTroubleshooting.pdf
HDF5 Dynamically Loaded Filters
The HDF5 library can use third-party dynamically loaded filter plugins which are used for forms of compression that are not built into the library itself. "Dynamically loaded" means that the filter plugins are not compiled into the HDF5 library but reside in separate library files. The HDF5 library looks for these plugins and, if it finds them, loads them, and their features become available. (For HDF5 experts, dynamically loaded filter plugins are described at https://support.hdfgroup.org/releases/hdf5/documentation/rfc/HDF5DynamicallyLoadedFilters.pdf.)
The default location where the HDF5 libraries look for filter plugins is:
%ALLUSERSPROFILE%/hdf5/lib/plugin (%ALLUSERSPROFILE% is C:\ProgramData on most systems)
The user can override the default locations by setting the HDF5_PLUGIN_PATH environment variable to the path to the user's plugins prior to launching Igor.
Igor Pro ships with the plugins provided by The HDF Group at https://www.hdfgroup.org/downloads/hdf5. These plugins include:
BLOSC, BLOSC2, BSHUF, BZ2, JPEG, LZ4, LZF, ZFP, and ZSTD
The plugins BLOSC2 and ZSTD are new in Igor Pro 10. Igor supports decoding datasets written with these filters. It does not yet support encoding with these filters.
The filters are shipped in "IgorBinaries_x64/hdf5plugins". When Igor starts, it tells the HDF5 library to look in these locations first for plugins, before looking in the default filter location or in the locations specified by the HDF5_PLUGIN_PATH environment variable if it exists. This means that the HDF5 library will find the filter plugins that ship with Igor, not those in other locations.
There are other "registered filtered plugins" besides the ones listed above. A comprehensive list can be found at https://github.com/HDFGroup/hdf5_plugins/blob/master/docs/RegisteredFilterPlugins.md. It is unlikely that you will need other filter plugins. If you do, you can put them in the default location or the location specified by the HDF5_PLUGIN_PATH environment variable. The HDF5 library will look for them there after not finding them in Igor's hdf5plugins folder.
HDF5 Dimension Scales
The HDF5 library supports dimension scales through the H5DS API. A dimension scale is a dataset that provides coordinates for another dataset. A dimension scale for dimension 0 of a dataset is analogous to an X wave in an Igor XY pair. The analogy applies to higher dimensions also.
Dimension scales are primarily of use in connection with the netCDF-4 format which is based on HDF5. Most Igor users do not need to know about dimension scales.
In the netCDF file format, the term "variable" is used like the term "dataset" in HDF5. Each variable of dimension n is associated with n named dimensions. Another variable, called a "coordinate variable", can supply values for the indices of a dimension, like an X wave supplies values for an XY pair in Igor.
The association between a variable and its coordinate variables is established when the variable is created. A given coordinate variable typically provides values for a given dimension of multiple variables. For example, a netCDF file may define coordinate variables named "latitude" and "longitude" and multiple 2D variables (images) whose X and Y dimensions are associated with "latitude" and "longitude".
The netCDF-4 file format is implemented using HDF5. The netCDF library uses the HDF5 dimension scale feature to implement coordinate variables and their associations with variables.
HDF5 Dimension Scale Support in Igor
The HDF5DimensionScale operation supports the creation and querying of HDF5 dimension scales. It was added in Igor Pro 9.00.
The operation is implemented using keyword=value syntax for setting parameters and simple keywords without values for invoking an action. For example, these commands convert a particular dataset into a dimension scale and then attach that dimension scale to another dataset:
// Convert a dataset named XScale into a dimension scale with name "X"
HDF5DimensionScale dataset={fileID,"XScale"}, dimName="X", setScale
// Attach dimension scale XScale to dimension 0 of dataset Data0
HDF5DimensionScale scale={fileID,"XScale"}, dataset={fileID,"Data0"}, dimIndex=0, attachScale
For details, see HDF5DimensionScale.
HDF5 Dimension Scale Implementation Details
If you view netCDF-4 files using Igor's HDF5 Browser, this section will help you to understand what you see in the browser.
The H5DS (dimension scale) API, which is part of the HDF5 "high-level" library, uses attributes to designate a dataset as a dimension scale, to associate a dimension scale with other datasets, and to keep track of the associations.
The most important attributes of a dimension scale are:
| CLASS | Set to "DIMENSION_SCALE" to indicate that the dataset is a dimension scale. | |
| NAME | Name of dimension such as "X". | |
| REFERENCE_LIST | An array of structures used to keep track of the datasets and dimensions to which the dimension scale is attached. Each element of the structure includes a reference to a dataset and a dimension index. | |
| The most important attributes of a dataset to which a dimension scale is attached are are: | ||
| DIMENSION_LIST | A variable-length array of references used to keep track of the dimensions used by each dimension of the dataset. The array has one column for each dataset dimension. Each column has one row for each dimension scale attached to the corresponding dataset dimension. A given dataset dimension can have multiple attached dimension scales. | |
| DIMENSION_LABELS | An 1D array containing labels for the dimensions of the dataset. | |
HDF5 Dimension Scale Reference
For experts, here are some sources of additional information on HDF5 dimension scales and related netCDF-4 features:
H5DS API Reference
https://support.hdfgroup.org/documentation/hdf5/latest/_h5_d_s__u_g.html
HDF5 Dimension Scale Specification (2005)
https://support.hdfgroup.org/documentation/hdf5-docs/hdf5_topics/H5DS_Spec.pdf
NetCDF Interoperability With HDF5
https://www.unidata.ucar.edu/software/netcdf/docs/interoperability_hdf5.html
https://www.unidata.ucar.edu/blogs/developer/en/entry/dimensions_scales
NetCDF FAQ
https://www.unidata.ucar.edu/software/netcdf/docs/faq.html
NetCDF Documentation
https://www.unidata.ucar.edu/software/netcdf/docs/index.html
NetCDF Data Model
https://www.unidata.ucar.edu/software/netcdf/docs/netcdf_data_model.html
HDF5/netCDF-4 Conventions
http://stcorp.github.io/harp/doc/html/conventions/hdf5.html
Other HDF5 Issues
This section is mostly of interest to advanced HDF5 users.
HDF5 String Formats
Strings can be written to HDF5 files as datasets or as attributes using several formats:
-
Variable length
-
Fixed length with null termination
-
Fixed length with null padding
-
Fixed length with space padding
In addition, strings can be marked as either ASCII or UTF-8.
Usually you do not need to know or care about the format used to write strings. However, some programs do not support all of the formats. If you attempt to load an HDF5 file written by Igor into one of those programs, you may get an error. In that event, you may be able to fix the problem by controlling the string format used to write the file. The rest of this section provides information that may help in that event.
The variable-length string format is most useful when writing a dataset or attribute containing multiple strings of different lengths. For example, when a dataset containing the two strings "ABC" and "DEFGHIJKLMNOP" is written as variable length, each string and its length is stored in the HDF5 file. That requires 3 bytes for "ABC" and 13 bytes for "DEFGHIJKLMNOP" plus the space required to store the length for each string. If these strings were written as fixed length, the fixed length for the dataset or attribute would have to be at least 13 bytes and at least 10 bytes would be wasted by padding when writing "ABC".
The fixed-length string format is most useful when writing a dataset or attribute consisting of a single string. For example, "ABC" can be written using a 3-byte fixed-length datatype with 0 padding bytes.
If more than one string is written as fixed length, the padding mode determines how extra bytes are filled. In null-terminated mode, the extra bytes are filled with null (0) and the first null marks the end of a string. In null-padded mode, the extra bytes are filled with null (0) all consecutive nulls at the end of the string mark the end of the string. In space-padded mode, the extra bytes are filled with space all consecutive spaces at the end of the string mark the end of the string.
In addition to the variable-length versus fixed-length issue, there is a text encoding issue with HDF5. A string dataset or attribute is marked as either ASCII or UTF-8 depending on the software that wrote it. The marking does not guarantee that the text is valid ASCII or valid UTF-8 as the HDF5 library does not check to make sure that written text is valid as marked nor does it do any text encoding conversions. The marking is merely a statement of the intended text encoding. Some software may fail when reading datasets or attributes marked as ASCII or UTF-8 because the HDF5 library does require that the reading program use a compatible datatype.
With some exceptions explained below, you can control the string format used to save datasets and attributes using the /STRF={fixedLength,paddingMode,charset} flag with the HDF5SaveData and HDF5SaveGroup operations. The /STRF flag was added in Igor Pro 9.00.
If fixedLength is 0, HDF5SaveData writes strings using a variable-length HDF5 string datatype. If fixedLength is greater than 0, HDF5SaveData writes strings using a fixed-length HDF5 string datatype of the specified length with padding specified by padding mode. If fixedLength is -1, HDF5SaveData determines the length of the longest string to be written for a given dataset or attribute and writes strings using a fixed-length HDF5 string datatype of that length with padding specified by paddingMode.
If paddingMode is 0, HDF5SaveData writes fixed-length strings as null terminated strings. If paddingMode is 1, HDF5SaveData writes fixed-length strings as null-padded strings. If paddingMode is 2, HDF5SaveData writes fixed-length strings as space-padded strings. When writing strings as variable length (fixedLength=0), paddingMode is ignored.
If charset is 0, HDF5SaveData writes strings marked as ASCII. If charset is 1, HDF5SaveData writes strings marked as UTF-8.
An exception is zero-length datasets or attributes which are always written as variable-length UTF-8. Another exception is string variables written by HDF5SaveGroup which are always written as fixed-length null padded UTF-8.
This table shows the default string format used for various situations if you omit /STRF and whether or not HDF5SaveData and HDF5SaveGroup honor the /STRF flag for the corresponding situation:
| HDF5SaveData Default | HDF5SaveData Behavior | |||
| Text Wave Zero Element as Dataset | Variable, NULLPAD, UTF-8 | Ignores /STRF | ||
| Text Wave Single Element as Dataset | Variable, NULLPAD, UTF-8 | Honors /STRF | ||
| Text Wave Multiple Elements as Dataset | Variable, NULLPAD, UTF-8 | Honors /STRF | ||
| String Variable | N/A | HDF5SaveData cannot save string variables | ||
| Text Wave Zero Element as Attribute | Variable, NULLPAD, UTF-8 | Ignores /STRF | ||
| Text Wave Single Element as Attribute | Fixed, NULLPAD, UTF-8 | Honors /STRF | ||
| Text Wave Multiple Elements as Attribute | Variable, NULLPAD, UTF-8 | Honors /STRF | ||
| HDF5SaveGroup Default | HDF5SaveGroup Behavior | |||
| Text Wave Zero Element as Dataset | Variable, NULLPAD, UTF-8 | Ignores /STRF | ||
| Text Wave Single Element as Dataset | Variable, NULLPAD, UTF-8 | Honors /STRF | ||
| Text Wave Multiple Elements as Dataset | Variable, NULLPAD, UTF-8 | Honors /STRF | ||
| Zero-Length String Variable | Variable, NULLPAD, UTF-8 | Ignores /STRF | ||
| String Variable > Zero-Length | Fixed, NULLPAD, UTF-8 | Ignores /STRF | ||
| Text Wave Zero Element as Attribute | N/A | HDF5SaveGroup cannot save attributes | ||
| Text Wave Single Element as Attribute | N/A | HDF5SaveGroup cannot save attributes | ||
| Text Wave Multiple Elements as Attribute | N/A | HDF5SaveGroup cannot save attributes | ||
HDF5 String Variable Text Encoding
Igor writes string variables as UTF-8 fixed-length string datasets.
String variables may contain null bytes and text that is invalid as UTF-8. This would occur, for example, if a variable were used to contain binary data. Such string variables are still written as UTF-8 fixed-length string datasets.
HDF5 Wave Text Encoding
For background information on wave text encoding, see Wave Text Encodings.
Igor text wave contents are written as variable-length string datasets using UTF-8 text encoding. Other wave elements (units, note, dimension labels) are written as UTF-8 fixed-length string attributes.
Text wave contents may contain null bytes and text that is invalid as UTF-8. This would occur, for example, if a text wave were used to contain binary data. Such contents are still written as UTF-8 variable-length string datasets.
Wave text elements can be marked in Igor as using any supported text encoding. No matter how a wave's text elements are marked, they are written to HDF5 as UTF-8 strings. Consequently, if you save a wave that uses non-UTF-8 text encodings to an HDF5 file and the load it back into Igor, its text encodings change but the characters represented by the text do not.
An exception applies to wave elements marked as binary (see Text Waves Containing Binary Data for background information). When you save a wave containing one or more binary elements as an HDF5 dataset, Igor adds the IGORWaveBinaryFlags attribute to the dataset. This attribute identifies the wave elements marked as binary using bits as defined for the WaveTextEncoding function. When you load the dataset from an HDF5 file, Igor restores the binary marking for the wave elements corresponding to the bits set in the attribute. The IGORWaveBinaryFlags attribute was added in Igor Pro 9.00.
In Igor Pro 9.00 and later, the HDF5LoadData and HDF5LoadGroup operations can check for binary data loaded into text waves from string datasets and mark such text waves as containing binary. If you load binary data from string datasets, see bit 1 of the /OPTS flag of those operations for details.
Igor Compatibility With HDF5
This section discusses issues relating to various HDF5 library versions.
You don't need to know this information unless you are using very old software based on HDF5 library versions earlier than 1.8.0.
The following table lists various Igor and HDF5 versions:
| Igor Version | HDF5 Library Version |
|---|---|
| Igor Pro 6.03 | 1.6.3 (released on 2004-09-22) |
| Igor Pro 6.10 to 6.37 | 1.8.2 (released on 2008-11-10) |
| Igor Pro 7.00 to 7.08 | 1.8.15 (released on 2015-05-04) |
| Igor Pro 8.00 to 8.04 | 1.10.1 (released on 2017-04-27) |
| Igor Pro 9.00 to 9.0x | 1.10.7 (released on 2020-09-15) |
| Igor Pro 10.00 | 1.14.5 (released on 2024-09-30) |
HDF5 Compatibility Terminology
For the purposes of this discussion, the term "old HDF5 version" means a version of the HDF5 library earlier than 1.8.0. The term "old HDF5 program" means a program that uses an old HDF5 version. The term "old HDF5 file" means an HDF5 file written by an old HDF5 program.
Reading Igor HDF5 Files With Old HDF5 Programs
By default, Igor Pro 10 and later create HDF5 files that work with programs compiled with HDF5 library version 1.8.0 or later. HDF5 1.8.0 was released in February of 2008. For most uses, this default compatibility will work fine.
The rest of this section is of interest only if you need to read Igor HDF5 files using very old HDF5 programs which use HDF5 library versions earlier than 1.8.0.
As explained at https://docs.hdfgroup.org/archive/support/HDF5/doc/ADGuide/CompatFormat180.html, software compiled with HDF5 1.6.2 (released on 2004-02-12) or before is incompatible with HDF5 files produced by HDF5 1.8.0 or later.
If you need to read files written by Igor Pro 10 and later using software that uses HDF5 1.6.3 (released on 2004-09-22) through 1.6.10 (released on 2009-11-10), you need to tell Igor to use compatible HDF5 formats. You do this by executing this command:
SetIgorOption HDF5LibVerLowBound=0
When you do this, you lose these features:
-
The ability to save attributes larger than 65,535 bytes ("large attributes")
You will get an error in the unlikely event that you attempt to write a large attribute to a group or dataset.
This includes saving a wave with a very large wave note or with a very large number of dimension labels via HDF5SaveData or HDF5SaveGroup or by saving an HDF5 packed experiment file. It also includes adding a large attribute using HDF5SaveData/A.
-
The ability to sort groups and datasets by creation order
This feature is provided by the HDF5 browser but is supported only for files in which it is enabled when the file is created.
-
The ability to write attributes so that they can be read in creation order
To restore Igor to normal operation, execute:
SetIgorOption HDF5LibVerLowBound=1
The effect of SetIgorOption lasts only until you restart Igor.
Typically, if you need to set HDF5LibVerLowBound at all, you would do this once at startup. Do not call SetIgorOption to set HDF5LibVerLowBound while an Igor preemptive thread is making HDF5 calls.
Writing to Old HDF5 Files
The HDF5 documentation does not spell out all of the myriad compatibility issues between various HDF5 library versions. The information in this section is based on our empirical testing.
See HDF5 Compatibility Terminology for a definition of terms used in this section.
If you use Igor to open an old HDF5 file and write a dataset to it, the new dataset is not readable by old HDF5 programs. Also, the group containing the new dataset cannot be listed by old HDF5 programs. In other words, writing to an old file in new format makes the old file at least partially unreadable by old software.
If you have old HDF5 files and you rely on old HDF5 programs to read them, open such files for read only, not for read/write. Also make sure the files are thoroughly backed up.
HDF5 Packed Experiment Files
In Igor Pro 9 and later, you can save an Igor experiment in an HDF5 file. The main advantage is that the data is immediately accessible to a wide array of programs that support HDF5. The main disadvantage is that you will need Igor Pro 9 or later to open the file in Igor. Also HDF5 is considerably slower than PXP for experiments with very large numbers of waves.
To save an experiment in HDF5 packed experiment format, choose File→Save Experiment As. In the resulting Save File dialog, choose "HDF5 Packed Experiment Files (*.h5xp)" from the pop-up menu under the list of files. Then click Save.
If you want to make HDF5 packed experiment format your default format for saving new experiments, choose Misc→Miscellaneous Settings. In the resulting dialog, click the Experiment category on the left. Then choose "HDF5 Packed (.h5xp)" from the Default Experiment Format pop-up menu.
For normal use you don't need to know the details of how Igor stores data in HDF5 packed experiment files, but, if you are curious, you can get a sense by opening such a file using the HDF5 Browser (Data→Load Waves→New HDF5 Browser).
The rest of this topic is for the benefit of programmers who want to read or write HDF5 packed experiment files from other programs. It is assumed that you are an experienced Igor and HDF5 user.
HDF5 Packed Experiment File Organization
An HDF5 packed experiment file has the following general organization:
/
History
History // Contents of the history area window
Packed Data
<Waves, variables and data folders here>
Shared Data
<Paths to shared wave files here>
Free Waves
<Free waves here>
Free Data Folders
<Free data folders here>
Packed Procedure Files
Procedure // Contents of the built-in procedure window
<Zero or more packed procedure files>
Shared Procedure Files
<Paths to shared procedure files>
Packed Notebooks
<Packed notebook files>
Shared Notebooks
<Paths to shared notebook files>
Symbolic Paths
<Paths to folders associated with symbolic paths>
Recreation
Recreation Procedures // Experiment recreation procedures
Miscellaneous
Dash Settings (dataset)
Recent Windows Settings (dataset)
Running XOPs (dataset)
Pictures
<Picture datasets here>
Page Setups
Page Setup for All Graphs (dataset)
Page Setup for All Tables (dataset)
Page Setup for Built-in Procedure Window (dataset)
<Datasets for page layout page setups here>
XOP Settings
<XOP settings datasets here>
Any of the top-level groups can be omitted if it is not needed. For example, if the experiment has no free waves, Igor writes no Free Waves group.
The Shared Data group contains datasets containing full paths to Igor binary wave (.ibw) files referenced by the experiment. The paths are expressed as Windows paths on Windows.
The Shared Procedure Files group contains datasets containing full paths to procedure (.ipf) files referenced by the experiment. The paths are expressed as Windows paths on Windows. Global procedure files and #included procedure files are not part of the expeirment and are not included.
The Shared Notebooks group contains datasets containing full paths to notebook (.ifn or any plain text file type) files referenced by the experiment. The paths are expressed as Windows paths on Windows.
The Symbolic Paths group includes only user-created symbolic paths, not the built-in symbolic paths Igor, IgorUserFiles, or home which by definition points to the folder containing the packed experiment file. The paths are expressed as Windows paths on Windows. If there are no user-created symbolic paths, Igor writes no Symbolic Paths group.
The Recreation Procedures dataset contains the experiment recreation procedures written by Igor to recreate the experiment.
The Miscellaneous group contains datasets and subgroups. Any of these objects can be omitted if not needed. The format of the data in the Miscellaneous group is subject to change and not documented. If your program reads or writes HDF5 packed experiment files, we recommend that you not attempt to read or write these objects.
HDF5 Packed Experiment File Hierarchy
An HDF5 file is a "directed graph" rather than a strict hierarchy. This means that it is possible to create strange relationships between objects, such as a group being a child of itself, or a dataset being a child of more than one group. Such strange relationships are not allowed for HDF5 packed experiment files - they must constitute a strict hierarchy.
Storage of Plain Text in HDF5 Packed Experiment Files
Several kinds of plain text items are stored in experiment files, including:
-
The contents of the built-in procedure window
-
The contents of the built-in history window
-
The contents of packed procedure files
-
The contents of packed plain text notebook files
-
Text data contents of text waves
-
Text properties of numeric waves (e.g., units, wave note, dimension labels)
-
String variables
Igor stores such text as strings (HDF5 class H5T_STRING) using UTF-8 text encoding (H5T_CSET_UTF8) when writing an HDF5 packed experiment file.
Occasionally an item that is expected to be valid UTF-8 will contain byte sequences that are invalid in UTF-8 or contain null bytes. Examples include text waves and string variables that are being used to carry binary rather than text data. Such data is still written as UTF-8 string datasets.
Plain Text Files
"Plain text files" in this context refers to history text, recreation procedures, procedure files, and plain text notebooks. Igor writes plain text files as UTF-8 string datasets.
Waves
Igor writes text waves as UTF-8 string datasets. For details, see HDF5 Wave Text Encoding.
String Variables
Igor writes string variables as UTF-8 fixed-length string datasets. For details, see HDF5 String Variable Text Encoding.
Writing HDF5 Packed Experiments
This section is for programmers who want to write HDF5 packed experiment files from programs other than Igor. This section assumes that you understand the information presented in the preceding sections of HDF5 Packed Experiment Files.
If you open an HDF5 packed experiment using the HDF5 Browser you will see that the file (top-level group displayed as "root" in the HDF5 file) has a number of attributes. The only required attribute is the IGORRequiredVersion attribute which specifies the minimum version of Igor required to open the experiment. Write 9.00 for this attribute, or a larger value if your HDF5 packed experiment requires a later version of Igor.
Waves in HDF5 Files
Igor writes waves to HDF5 files as datasets with attributes representing wave properties. For example, the IGORWaveType attribute represents the data type of the wave - see WaveType for a list of data types. See the discussion of the /IGOR flag of the HDF5SaveData operation for a list of HDF5 wave attributes.
Wave Reference Waves and Data Folder Reference Waves
A wave reference wave (see Wave Reference Waves), or "wave wave" for short, is a wave whose elements are references to other waves. Each wave referenced by a wave wave has an IGORWaveID attribute that specifies the wave ID of the referenced wave. The contents of each element of a wave wave is the wave ID for the referenced wave. A wave ID of zero indicates a null wave reference. On loading the experiment, Igor restore each element of each wave wave so that it points to the appropriate wave.
A data folder reference wave (see Data Folder Reference Waves), or "DFREF wave" for short, is a wave whose elements are references to data folders. Each data folder referenced by a DFREF wave has an IGORDataFolderID attribute that specifies the data folder ID of the referenced data folder. The contents of each element of a DFREF wave is the data folder ID for the referenced data folder. A data folder ID of zero indicates a null data folder reference. On loading the experiment, Igor restore each element of each DFREF wave so that it points to the appropriate data folder.
Writing and restoring wave waves and DFREF waves is complicated and tricky. If your program writes HDF5 packed experiment files, we recommend that you not attempt to write these objects. If your program loads HDF5 packed experiment files, we recommend that you ignore ignore wave waves (IGORWaveType=16384) and DFREF waves (IGORWaveType=256).
Free Waves and Free Data Folders
Igor writes representations of Free Waves and Free Data Folders to HDF5 packed experiment files and restores free waves and free data folders when the experiment is loaded.
Each free wave is written as a dataset in the Free Waves group. For a free wave to exist, it must be referenced by a wave wave in the experiment.
Each free root data folder and its descendents are written as a group and subgroups in the Free Data Folders group. For a free root data folder to exist, it must be referenced by a DFREF wave in the experiment.
Writing and restoring free waves and free data folders is a complicated and tricky. If your program writes HDF5 packed experiment files, we recommend that you not attempt to write these objects. If your program loads HDF5 packed experiment files, we recommend that you ignore the Free Waves and Free Data Folders groups.
HDF5 Packed Experiment Issues
In rare cases, Igor experiments cannot be written in HDF5 packed format because of name conflicts. For details, see Object Name Conflicts and HDF5 Files.